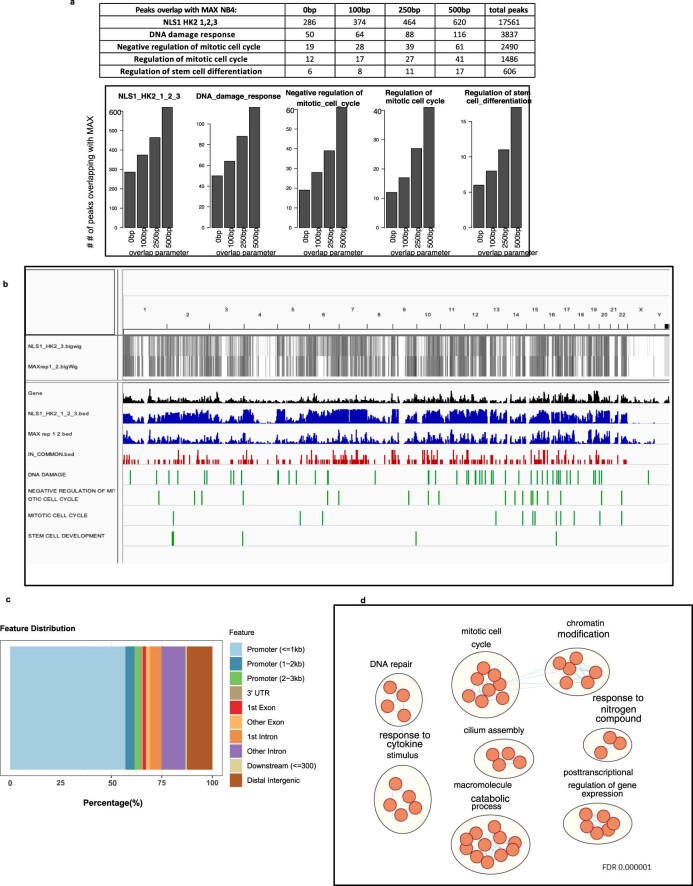
Extended Data Fig. 8. Comparative analysis of nuclear HK2 and MAX ChIP-seq.
(a) Number of overlapping peaks between NLS1 HK2 and MAX ChIP–seq peaks using different overlap parameters. (b) Visualization of overlapping peaks between NLS1-HK2 and MAX ChIP–seq peaks in NB4 cells on a full genome view (0bp parameter). (c) Distribution of the peaks in common between NLS1-HK2 and MAX (0bp parameter). (d) Pathways (GO BP) enriched in the peaks common between NLS1-HK2 and MAX (0bp parameter), at FDR <0.000001 using the binomial test.