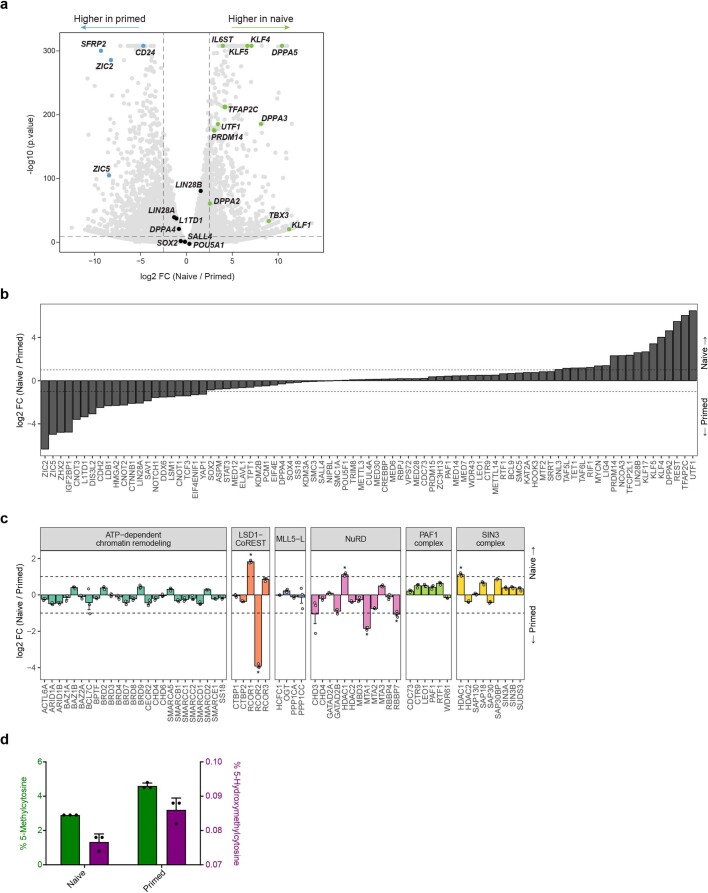
Extended Data Fig. 1. Global analysis of chromatin proteome and transcriptome in naive and primed hPSCs.
a. Volcano plot showing differential gene expression as detected by RNA-seq between naive and primed hPSCs (n = 3 biologically independent samples). Core pluripotency factors (black) as well as factors specific to each pluripotent state (blue for primed; green for naive) are highlighted. Dashed lines indicate p-value < 0.05 and log2fold change > 2.5 (two-sided student’s t test). P-values can be found in Supplementary Table 8. b. Chromatin occupancy of all proteins with a potential role in stem cell maintenance in naive and primed hPSCs (n = 3 biologically independent samples). Protein names were collected from AmiGO (http://amigo.geneontology.org/; ‘stem cell maintenance’ GO:0019827)38,50. The following pluripotency-associated proteins were not detected in our dataset: ASCL2, BMP7, BMPR1A, DAZL, DLL1, ERAS, ESRRB, FANCC, FGF10, FGF4, FGFR1, FOXO3, FZD7, HES1, HES5, HESX1, ID1, ID2, ID3, JAG1, KIT, KLF10, KLF2, LBH, LDB2, LIF, LOXL2, LRP5, MCPH1, MED21, MED27, MMP24, MYC, NANOG, NANOS2, NODAL, NOG, NOTCH2, NR0B1, NR2E1, PADI4, PAX2, PAX8, PELO, PHF19, PIWIL2, PRDM16, PROX1, PRRX1, PTN, RAF1, SETD6, SFPI1, SFRP1, SIX2, SKI, SMO, SOX9, SPI1, TAL1, TBX3, TCF15, TCL1, TERT, TP63, TUT4, WNT7A, WNT9B, ZFP36L2, ZNF322, ZNF358, ZNF706. c. ChEP analysis of differential abundance of chromatin-associated complexes in naive and primed hPSCs (n = 3 biologically independent samples), supplementing Fig. 1d,e. Data are presented as mean values + /- SEM. Dashed lines represent 2-fold change. Asterisks indicate p-value < 0. 05 and fold change > 2 (two-sided student’s t test). Low-change proteins (log2 FC < 0.5) involved in ATP-dependent chromatin remodelling (Fig. 1e) are shown. d. Mass spectrometry analyses of global levels of DNA methylation (green) and DNA hydroxymethylation (purple) in naive and primed hPSCs (n = 3 biologically independent samples) Data are presented as mean values + /- SD. Underlying source data is provided in Source Data Extended Data Fig. 1.