Abstract
Iron is essential for many biological processes, but iron levels must be tightly regulated to avoid harmful effects of both iron deficiency and overload. Here, we perform genome-wide association studies on four iron-related biomarkers (serum iron, serum ferritin, transferrin saturation, total iron-binding capacity) in the Trøndelag Health Study (HUNT), the Michigan Genomics Initiative (MGI), and the SardiNIA study, followed by their meta-analysis with publicly available summary statistics, analyzing up to 257,953 individuals. We identify 123 genetic loci associated with iron traits. Among 19 novel protein-altering variants, we observe a rare missense variant (rs367731784) in HUNT, which suggests a role for DNAJC13 in transferrin recycling. We further validate recently published results using genetic risk scores for each biomarker in HUNT (6% variance in serum iron explained) and present linear and non-linear Mendelian randomization analyses of the traits on all-cause mortality. We find evidence of a harmful effect of increased serum iron and transferrin saturation in linear analyses that estimate population-averaged effects. However, there was weak evidence of a protective effect of increasing serum iron at the very low end of its distribution. Our findings contribute to our understanding of the genes affecting iron status and its consequences on human health.
Subject terms: Biomarkers, Genome-wide association studies
A GWAS on the four major iron-related biomarkers is conducted using data from the Trøndelag Health Study (HUNT), Michigan Genomics Initiative (MGI), and SardiNIA study, to identify 123 loci associated with iron homeostasis and effect on all-cause mortality.
Introduction
Iron is essential for a variety of physiological processes in the human body, but excess iron is toxic. Iron overload is associated with a wide range of health problems, including liver damage, type 2 diabetes, cardiovascular disease, and neurodegenerative diseases such as Alzheimer’s disease1–3, while long-term iron deficiency causes anemia, which can disrupt cognitive function and the immune system4–6. Because of the damaging effects of both deficiency and overload, iron metabolism is tightly regulated7.
Iron is bound, transported, and delivered around the body by the transferrin glycoprotein8, while the main intracellular iron storage, ferritin, provides a long-term reserve of iron for the formation of hemoglobin and other heme proteins9–11. Serum iron, serum ferritin, transferrin saturation percentage (TSP), and the total iron-binding capacity (TIBC) of transferrin are biochemical measurements that are commonly used together to assess an individual’s iron status12. Both high and low TSP have been associated with an increased mortality risk in observational studies13–16, although the underlying pathophysiologic mechanisms are unclear.
Mutations in various iron metabolism genes can cause both iron deficiency and overload17–19. Genetic variants in the transferrin gene, TF, and in the homeostatic iron regulator gene, HFE, have been estimated to account for about 40% of genetic variation in transferrin levels20. A recent genome-wide association study (GWAS) meta-analysis21 of serum iron, ferritin, TSP, and TIBC from Iceland, UK, and Denmark reported 46 novel loci associated with at least one of these biomarkers, implicating proteins involved in iron homeostasis. Identifying additional genetic loci associated with iron status could further increase our understanding of pathophysiologic mechanisms underlying dysregulated iron levels. Furthermore, genetic variants from the most recent study21 could improve existing genetic risk scores (GRS) that have been widely used to assess the causal associations of iron status on a range of outcomes using Mendelian Randomization22–27 (MR). However, the new GRSs have not yet been validated in an independent study. Further, despite the observed damaging effects of both very high and very low iron stores, no previous MR studies have investigated the shape of the associations between genetically proxied iron status biomarkers and mortality. By validating the most recent genetic risk scores and using MR in an independent study (HUNT), we provide robust and novel insights into the causal associations between iron status biomarkers and all-cause mortality, particularly regarding non-linear relationships.
Here, we identify 123 loci associated with iron status, by combining three approaches: (i) genome-wide association studies of variants deeply imputed from the TOPMed reference panel28 in the Trøndelag Health Study (HUNT)29 and the Michigan Genomics Initiative (MGI), as well as variants imputed from a cohort-specific reference panel in SardiNIA30 (ii) association tests with genotyped coding variants selected from low-coverage (5×) whole-genome sequencing in HUNT, (iii) genome-wide meta-analyses of HUNT, MGI, SardiNIA and summary statistics from deCODE, Interval and the Danish Blood Donor Study (DBDS)21. The analyses included up to 257,953 individuals (57% females, 43% males) with measured iron status biomarkers. We evaluate the variance explained by previously published variants for serum iron, serum ferritin, TSP and TIBC in HUNT. Furthermore, we use the genetic variants for the iron status biomarkers to estimate the average causal effect of a population shift in the biomarker distributions on all-cause mortality (the population-averaged effect), and investigate the shape of the causal relationships using non-linear MR. We find evidence of an average harmful effect of increasing serum iron and transferrin saturation in the general population, but also weak evidence of a protective effect of increasing serum iron at the very low end of its distribution.
Results
Discovery of genetic loci associated with iron status
We identified 123 genetic loci associated (p-value <5 × 10−8) with at least one iron related biomarker (Tables 1–2, Supplementary Data 1, Supplementary Figs. 1–4, and Supplementary Note 1) in genome-wide association meta-analyses of the iron status biomarkers in six cohorts: HUNT, MGI, SardiNIA, deCODE, Interval and DBDS (Supplementary Data 2). To the best of our knowledge, 66 of these loci were novel for any iron status biomarker, while 57 had been reported for at least one of the biomarkers in previous studies. Additionally, among the 57 known iron status loci, 12 loci were for the first time associated with one or more additional biomarkers, further emphasizing the role of these loci in iron-related biological processes. Among the 62 unique index (lowest p-value) variants in novel iron status loci that had been imputed in more than one study, 49 had consistent directions of effects across all the analyzed studies. We also identified three novel missense variants associated with at least one biomarker among the variants ascertained from low-pass whole genome sequencing and selected for targeted genotyping in HUNT (Supplementary Data 3).
Table 1.
Novel loci associated with the iron status biomarkers serum iron, TSP, and TIBC in a genome-wide association meta-analysis.
| Trait | rsID-Alt | Effect | SE | P-value | MAF | Nearest gene(s) | Consequence | N | Studies |
|---|---|---|---|---|---|---|---|---|---|
| TIBC | rs514595-C | 0.03 | 0.005 | 6.5E−10 | 0.155 | AKR1A1;NASP | 208,422 | H;M;S;dC;I | |
|
TIBC S-Iron |
rs137891701-D rs61804206-G |
0.04 0.03 |
0.006 0.005 |
4.9E−9 4.7E−10 |
0.129 0.142 |
SDHC;FCGR2A RPL3P1P11;FCRLA |
R2 = 1.00 w/missense |
202,496 236,612 |
H;M;dC;I H;M;S;dC;I |
| TIBC | rs1435167-A | −0.02 | 0.004 | 1.6E−10 | 0.373 | EGLN1 | R2 = 0.89 w/missense | 208,422 | H;M;S;dC;I |
| S-Iron | rs7580634-T | −0.02 | 0.003 | 4.7E−10 | 0.464 | IL1F10;IL1RN | 236,612 | H;M;S;dC;I | |
| TIBC | rs11900910-T | 0.02 | 0.004 | 2.1E−11 | 0.451 | EPAS1 | 208,422 | H;M;S;dC;I | |
| TIBC | rs1861408-T | 0.02 | 0.004 | 7.8E−10 | 0.297 | SERTAD2;LINC01800 | 208,422 | H;M;S;dC;I | |
| TIBC | rs715-C | 0.02 | 0.004 | 4.5E−10 | 0.285 | CPS1 | R2 = 0.92 w/missense | 202,496 | H;M;S;dC;I |
| TIBC | rs781168893-T | −0.75 | 0.064 | 2.1E−31 | 0.003 | SLC12A8;ZNF148 | 56,667 | H | |
| S-Iron | rs571559677-C | −0.35 | 0.063 | 3.1E−8 | 0.002 | CLSTN2 | 67,171 | H;M | |
| TIBC | rs760897340-G | −0.97 | 0.104 | 7.5E−21 | 0.001 | DIPK2A;LNCSRLR | 56,667 | H | |
| TIBC | rs969672878-A | −0.90 | 0.103 | 2.3E−18 | 0.001 | LOC440982;LINC02032 | 56,667 | H | |
| TSP | rs218264-T | 0.02 | 0.004 | 4.2E−8 | 0.250 | LINC02283;LINC02260 | 198,516 | H;M;dC;I | |
| TIBC | rs185240714-A | −0.07 | 0.009 | 2.4E−13 | 0.040 | PELO | 204,433 | H;M;dC;I | |
| TIBC | rs41274050-T | 0.13 | 0.022 | 4.8E−9 | 0.008 | A1CF | 202,496 | H;M;dC;I | |
|
S-Iron TIBC |
rs10740134-C rs10740134-C |
0.02 0.02 |
0.003 0.004 |
7.8E−11 8.9E−10 |
0.487 0.487 |
REEP3 REEP3 |
R2 = 0.96 w/missense R2 = 0.96 w/missense |
236,612 198,516 |
H;M;S;dC;I H;M;dC;I |
| S-Iron | rs59213222-D | −0.03 | 0.005 | 9.3E−9 | 0.223 | ZMIZ1 | 230,682 | H;M;S;dC;I | |
| TIBC | rs143878994-A | 0.05 | 0.009 | 3.6E−8 | 0.046 | ARHGAP20 | 202,496 | H;M;S;dC;I | |
| TIBC | rs76895963-G | −0.08 | 0.015 | 4.2E−8 | 0.017 | CCND2-AS1 | 202,496 | H;M;S;dC;I | |
| TIBC | rs3782735-A | 0.02 | 0.004 | 2.4E−9 | 0.402 | LAG3 | 202,496 | H;M;S;dC;I | |
|
S-Iron TSP |
rs2900478-A rs2900478-A |
−0.02 −0.03 |
0.004 0.005 |
3.9E−8 1.4E−10 |
0.178 0.179 |
SLCO1B1 SLCO1B1 |
R2 = 0.90 w/missense R2 = 0.90 w/missense |
236,612 198,516 |
H;M;S;dC;I H;M;dC;I |
| TIBC | rs1150975-A | 0.02 | 0.004 | 4.1E−8 | 0.347 | LINC02422;RESF1 | 202,496 | H;M;S;dC;I | |
| TIBC | rs77371258-A | 0.04 | 0.006 | 3.5E−13 | 0.099 | POP5;CABP1 | R2 = 0.88 w/missense | 202,496 | H;M;S;dC;I |
| S-Iron | rs4759827-T | −0.02 | 0.004 | 1.5E−8 | 0.279 | ADGRD1 | 236,612 | H;M;S;dC;I | |
|
S-Iron TSP |
rs1958078-C rs1958078-C |
0.04 0.04 |
0.005 0.005 |
1.0E−14 1.7E−11 |
0.135 0.131 |
SMOC1 SMOC1 |
236,612 198,516 |
H;M;S;dC;I H;M;dC;I |
|
| TIBC | rs12879801-G | 0.03 | 0.004 | 1.5E−17 | 0.452 | ADAM21P1;SYNJ2BP-COX16 | 202,496 | H;M;S;dC;I | |
| TIBC | rs11159099-G | −0.03 | 0.004 | 5.2E−16 | 0.482 | YLPM1 | 202,496 | H;M;S;dC;I | |
| TIBC | rs8062982-C | 0.03 | 0.004 | 6.7E−12 | 0.401 | CHD9;LOC643802 | 202,496 | H;M;S;dC;I | |
| TIBC | rs34682685-A | 0.04 | 0.006 | 1.2E−12 | 0.100 | HPR | 202,496 | H;M;S;dC;I | |
| TIBC | rs2287322-G | 0.03 | 0.004 | 8.9E−13 | 0.239 | WDR81 | R2 = 0.97 w/missense | 202,496 | H;M;S;dC;I |
| TIBC | rs1292072-G | 0.03 | 0.004 | 5.0E−11 | 0.224 | VMP1;TUBD1 | 202,496 | H;M;S;dC;I | |
| TIBC | rs34284056-A | −0.02 | 0.004 | 1.6E−8 | 0.264 | ZCCHC2 | 202,496 | H;M;S;dC;I | |
| TIBC | rs111234557-G | −0.03 | 0.006 | 4.0E−9 | 0.121 | MAU2 | 202,496 | H;M;S;dC;I | |
| TIBC | rs12976652-C | −0.05 | 0.006 | 6.5E−18 | 0.105 | MYPOP | 202,496 | H;M;S;dC;I | |
|
S-Iron TIBC |
rs1800961-T rs1800961-T |
0.04 0.05 |
0.008 0.009 |
2.4E−8 4.0E−9 |
0.047 0.047 |
HNF4A HNF4A |
Missense Missense |
236,612 202,496 |
H;M;S;dC;I H;M;S;dC;I |
|
S-Iron TSP |
rs3747207-A rs3747207-A |
0.03 0.02 |
0.004 0.004 |
1.7E−12 1.1E−8 |
0.234 0.233 |
PNPLA3 PNPLA3 |
R2 = 1.00 w/missense R2 = 1.00 w/missense |
236,612 198,516 |
H;M;S;dC,I H;M;dC;I |
Associated traits: Serum iron (S-iron), Transferrin Saturation Percentage (TSP), Total Iron Binding Capacity (TIBC). Locus index variant with alternate allele/indel (rsID-Alt, D = deletion) per associated trait. Effect size and direction (Effect) with standard error (SE) and functional consequence (correlation R2 reported if index variant is strongly correlated (R2 > 0.8) with a missense variant.) is given for the alternate variant. Minor allele frequency (MAF), sample size (N), studies: HUNT (H), MGI (M), deCODE (dC), Interval (I), SardiNIA (S), DBDS (D). See Supplementary Data 1 for further details.
Table 2.
Novel loci associated with the iron status biomarker ferritin in a genome-wide association meta-analysis.
| rsID-Alt | Effect | SE | P-value | MAF | Nearest gene(s) | Consequence | N | Studies |
|---|---|---|---|---|---|---|---|---|
| rs477190-A | −0.02 | 0.003 | 1.3E−08 | 0.404 | FAM43B;CDA | 257,953 | H;M;dC;I;D | |
| rs604126-G | 0.02 | 0.004 | 2.7E−08 | 0.314 | BCL2L11;MIR4435-2HG | 257,953 | H;M;dC;I;D | |
| rs17050272-A | 0.02 | 0.003 | 1.1E−09 | 0.417 | LINC01101;GLI2 | 257,953 | H;M;dC;I;D | |
| rs17707216-A | 0.04 | 0.007 | 2.2E−08 | 0.076 | ZSWIM2;CALCRL | 257,953 | H;M;dC;I;D | |
| rs553656123-A | 0.11 | 0.020 | 1.1E−08 | 0.008 | NBEAL2 | 257,953 | H;M;dC;I;D | |
| rs142350264-I | −0.04 | 0.007 | 1.7E−09 | 0.098 | BSN | 246,139 | dC;I;D | |
| rs6822746-A | −0.02 | 0.004 | 6.3E−09 | 0.317 | SRD5A3-AS1;TMEM165 | 257,953 | H;M;dC;I;D | |
| rs4865796-A | 0.02 | 0.004 | 6.4E−09 | 0.319 | ARL15 | 257,953 | H;M;dC;I;D | |
| rs79694859-A | 0.05 | 0.009 | 3.2E−09 | 0.046 | PDE4D | 257,953 | H;M;dC;I;D | |
| rs35107257-A | 0.04 | 0.008 | 1.1E−08 | 0.058 | OCLN | 257,953 | H;M;dC;I;D | |
| rs970079-G | −0.02 | 0.003 | 7.0E−09 | 0.463 | RGMB | 257,953 | H;M;dC;I;D | |
| rs35486885-G | −0.06 | 0.008 | 5.2E−12 | 0.038 | HLA-DQB2;HLA-DOB | 257,953 | H;M;dC;I;D | |
| rs13215052-A | −0.03 | 0.005 | 5.7E−09 | 0.111 | TFEB | 257,953 | H;M;dC;I;D | |
| rs189899297-A | 0.02 | 0.004 | 4.7E−08 | 0.263 | ATP6V0D2;SLC7A13 | 246,139 | dC;I;D | |
| rs681099-C | −0.02 | 0.003 | 1.7E−08 | 0.455 | EIF3E | 255,619 | M;dC;I;D | |
| rs72775768-T | −0.02 | 0.004 | 4.2E−09 | 0.259 | INPP5E | 257,953 | H;M;dC;I;D | |
| rs9423600-G | 0.02 | 0.004 | 4.8E−08 | 0.268 | LINC02561;UCN3 | 257,953 | H;M;dC;I;D | |
| rs704017-G | −0.02 | 0.003 | 2.6E-08 | 0.378 | ZMIZ1-AS1 | 257,953 | H;M;dC;I;D | |
| rs17112021-G | 0.02 | 0.004 | 2.0E−10 | 0.275 | NKX2-3;SLC25A28 | 257,953 | H;M;dC;I;D | |
| rs7102016-T | 0.02 | 0.003 | 3.4E−08 | 0.444 | SBF2 | 257,953 | H;M;dC;I;D | |
| rs10750215-T | 0.02 | 0.004 | 1.5E-08 | 0.417 | MIR100HG;UBASH3B | 257,953 | H;M;dC;I;D | |
| rs9512463-C | −0.02 | 0.004 | 4.8E−09 | 0.226 | GPR12;USP12 | 257,953 | H;M;dC;I;D | |
| rs28715334-T | 0.02 | 0.004 | 6.4E−09 | 0.203 | SLC7A8 | 257,953 | H;M;dC;I;D | |
| rs764195359-G | −0.30 | 0.051 | 2.6E−09 | 0.002 | MGA | 255,619 | M;dC;I;D | |
| rs11634990-C | −0.03 | 0.005 | 2.0E−09 | 0.170 | IREB2 | 257,953 | H;M;dC;I;D | |
| rs62074125-C | 0.02 | 0.004 | 3.8E−08 | 0.288 | WNT3 | 257,953 | H;M;dC;I;D | |
| rs2952290-A | −0.02 | 0.004 | 1.9E−08 | 0.214 | PRKAR1A | R2 = 0.84 w/missense | 257,953 | H;M;dC;I;D |
| rs141253118-A | −0.12 | 0.023 | 4.6E−08 | 0.005 | NOTCH3;EPHX3 | 255,619 | M;dC;I;D | |
| rs2595586-G | −0.02 | 0.003 | 2.2E−08 | 0.414 | ATRN | 257,953 | H;M;dC;I;D | |
| rs6088374-C | −0.04 | 0.005 | 1.9E−16 | 0.169 | LINC00028;HM13 | 255,619 | M;dC;I;D | |
| rs540828131-G | −0.16 | 0.026 | 1.3E−09 | 0.007 | HMOX1 | 255,619 | M;dC;I;D |
Locus index variants with alternate allele/indel (rsID-Alt, I = insertion). Effect size and direction (Effect) with standard error (SE) and functional consequence (correlation R2 reported if index variant is strongly correlated (R2 > 0.8) with a missense variant) given for the alternate variant. Minor allele frequency (MAF), sample size (N), studies: HUNT (H), MGI (M), deCODE (dC), Interval (I), SardiNIA (S), DBDS (D). See Supplementary Data 1 for further details.
Genes in several associated loci coded for proteins with established functions in iron homeostasis (TF [transferrin]) SLC25A37 [mitoferrin-1], SLC25A28 [mitoferrin-2], SLC11A2 [divalent metal-transporter 1] and SLC40A1 [ferroportin-1], HFE [homeostatic iron regulator], TFRC [transferrin receptor], TFR2 [transferrin receptor 2], HAMP [hepcidin], ERFE [erythroferrone], HMOX1 [heme oxygenase], IREB2 [iron responsive element binding protein 2], EPAS1 [endothelial PAS Domain Protein 1] and TMPRSS6 [transmembrane serine protease 6])7,31–33 Four of the loci that had not been reported for iron status biomarkers in previous GWAS studies, contained known iron related genes (SLC25A28, HMOX1, IREB2, and EPAS1), providing additional confidence in the reported associations. With exception of HAMP and TFR2, these genes were the nearest gene to the index variant in the locus.
Protein-altering variants in meta-analysis loci
We identified 32 protein-altering single nucleotide polymorphisms (SNPs) in the meta-analysis, which were either index variants or variants in strong linkage disequilibrium (LD) (R2 > 0.8 or D’ = 1.0) with an index variant (Supplementary Data 1). In addition to SNPs known to be related to diseases such as hemochromatosis, atransferrinemia or iron deficiency anemia19,34–38, and variants that had previously been reported for at least one of the iron traits21,39, we identified 11 protein-altering variants in novel iron status loci (Supplementary Data 4): rs9427398 (FCGR2A), rs2437150 (SPRTN), rs1047891 (CPS1), rs41274050 (A1CF), rs1935 (JMJD1C), rs3742049 (COQ5), rs4149056 (SLCO1B1), rs2070863 (SERPINF2), rs883541 (WIPI1), rs1800961 (HNF4A) and rs738409 (PNPLA3). In known iron status loci, we further identified eight protein-altering variants not previously reported for any of the analyzed iron traits: rs367731784 (DNAJC13), rs3812594 (SEC16A), rs34376913 (C9orf163), rs445520 (SLC11A2), rs28929474 (SERPINA1), rs737700 (C16orf71), rs77542162 (ABCA6) and rs34654230 (RCN3).
Custom genotyped variants in HUNT: Protein-altering variants in iron status loci
Among the targeted candidate variants in HUNT identified by sequencing and clinical studies, we identified three additional, novel protein-altering variants (Supplementary Data 3) that were not included in the meta-analyses, and which were associated with iron status biomarkers. These were located in NRM (rs374815811), HLA-DRB5 (rs701884), and TFR2 (chr7:100629337:A:T, GRCh38).
Heritability and genetic correlation of iron status markers
We estimated the respective narrow-sense SNP heritability (variance explained, Vg/Vp ± 1 standard error) of serum iron (0.15 ± 0.01), TIBC (0.43 ± 0.01) and TSP (0.21 ± 0.01) in HUNT using Genome-wide Complex Trait Analysis (GCTA)40. We found pair-wise genetic correlations between 11% and 75% (Supplementary Data 5) for the four iron status biomarkers using LD Score Regression41 with the meta-analysis summary statistics. The TSP phenotype was derived from the serum iron and TIBC measurements, giving rise to the two strongest genetic correlations. The weakest correlation (iron vs TIBC) did not reach nominal significance (p-value = 0.35).
Functional mapping
We used Bayesian colocalization analysis to identify 86 unique pairs of GWAS loci and cis-expression quantitative trait locus (cis-eQTL) signals that showed sufficient overlap in at least one tissue to be consistent with a shared causal variant for the gene expression and the iron status biomarker (Supplementary Data 6). We found associations in a range of tissues which highlighted genes with established roles in iron metabolism (TF [posterior probability of a common causal variant, PP4 = 0.96], TMPRSS6 [PP4 = 0.82], ERFE [PP4 = 0.97–0.98], IREB2 [PP4 = 0.80], SLC40A1 [PP4 = 0.79–0.96])17,31. Additionally, our results confirmed previously reported genes (DUOX2 [PP4 = 0.76], HBS1L [PP4 = 0.98], IL6R [PP4 = 0.81–0.82], SLC25A37 [PP4 = 0.85], ABO [PP4 = 0.97], RNF43 [PP4 = 0.99])21,39, and identified novel genes interacting with previously reported genes, for example DUOXA242. Several iron status loci were also colocalized with cis-eQTL signals for genes in the major histocompatibility complex (MHC) other than HFE43, as well as with transcription regulators44–46, additional transporter proteins47,48 and transferases49,50.
Using Data-driven Expression Prioritized Integration for Complex Traits (DEPICT)51 we found an enriched (false discovery rate [FDR] < 0.05) expression of ferritin associated genes in the urogenital system, digestive system, and the hemic and immune system (Supplementary Data 7). Serum iron, TSP, and TIBC associated genes were not enriched in any tissue types at FDR < 0.05, however, the strongest enrichment for genes in all three traits were found in liver tissue, and particularly in hepatocytes (TSP, TIBC). The top ten genes per trait when prioritized based on similarity between the associated (p-value < 5 × 10−8) loci, included known iron regulatory genes (TFR2, HAMP, TFRC, and SLC40A1), genes in which we had identified protein-altering variants (IL6R, F5, GCKR, DUOX2, SERPINA1, ABCA6, and SLCO1B1), genes we found in the colocalization analysis (DUOXA2, IL1RN, and SLC25A37), as well as genes predicted to have iron ion binding and heme binding properties in gene ontology analyses (CYP3A43, CYP3A5)52 (Supplementary Data 8). Finally, we used DEPICT and found gene sets enriched with iron status associated genes (Supplementary Data 9). All the top ten gene sets enriched with iron associated loci and seven with TSP associated loci reached FDR < 0.05: One iron associated gene set, and five TSP associated gene sets were related to the liver (including abnormal liver physiology and gene sets related to metabolic processes), but also to inflammation (acute-phase response, decreased leukocyte cell number), coagulation (coagulation factor protein-protein interaction networks) and neurodevelopment (abnormal myelination). The top ten gene sets enriched with ferritin and TIBC associated loci included decreased circulating iron levels, decreased spleen iron levels, gene sets related to red blood cells (decreased hemoglobin, decreased hematocrit, erythrocyte homeostasis, and differentiation), as well as liver fibrosis and liver inflammation. However, these gene sets did not reach FDR < 0.05. We identified the 1% top-ranked genes per trait based on both physical distance to the associated genetic variants and functional similarity to other associated genes (Supplementary Data 10–13) using Polygenic Priority Scores (PoPS)53. The prioritized genes included the main known iron regulatory genes, several genes that were nearest to the meta-analysis index variants, and novel genes in which we identified protein-altering variants (WIPI1, SERPINF2, and HNF4A), further supporting a role for these genes in iron biology.
Phenome-wide association study (PheWAS) of biomarker loci
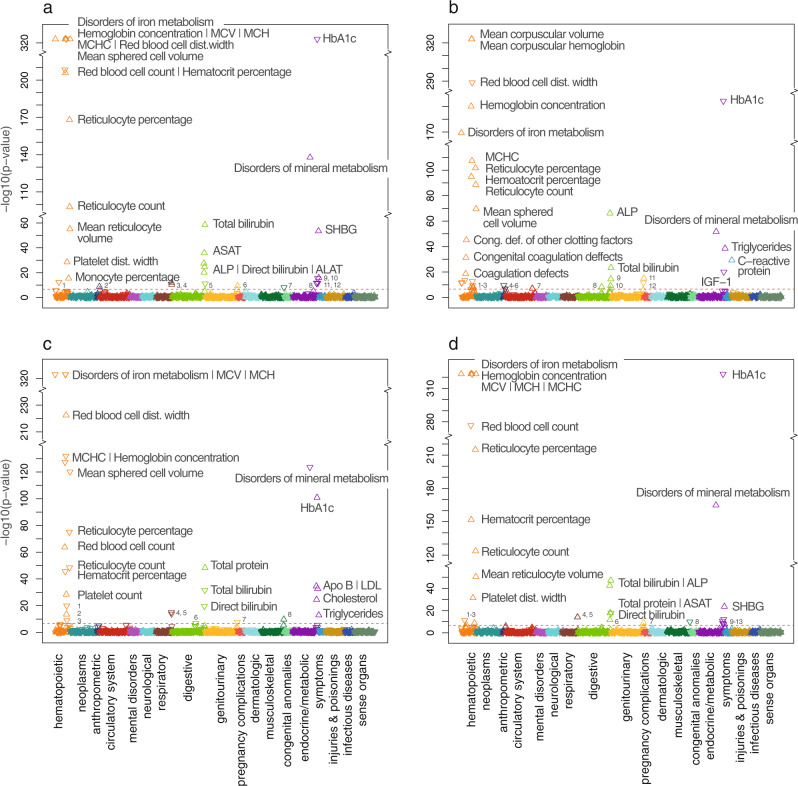
In total, 105 of the meta-analysis index variants were significantly associated (p-value < 2.3 × 10−7) with at least one additional disease phenotype (‘phecode’54), blood biomarker, or continuous trait in the UK Biobank. Of the 1473 phenotypes tested, 129 were significantly associated with at least one variant (Supplementary Data 14–19). The associations spanned numerous biological domains, but most associations were within the hematopoietic (615 variant-trait associations), endocrine/metabolic (303 variant-trait associations), and digestive (243 variant-trait associations) domains. The strongest (p-values < 1 × 10−300) and most numerous associations were seen for red blood cell-related traits (hemoglobin concentration, mean corpuscular hemoglobin, mean corpuscular volume, red blood cell distribution width, red blood cell count, hematocrit percentage, mean reticulocyte volume), where we found associations with variants in HFE (rs1800562, rs144861591, and rs79220007), TMPRSS6 (rs855791, rs2076085), HBS1L;MYB (rs9399136, rs56293029), LINC002283;LINC002260 (rs218264), HK1 (rs17476364) and IKZF1 (rs12718598). The strongest associations also included associations between ‘disorders of iron metabolism’ and the HFE variants (rs1800562, rs144861591 and rs79220007), platelet related traits (platelet count, platelet crit, mean platelet volume) and variants in HBS1L;MYB (rs9399136, rs56293029) and REEP3 (rs10740134), direct and total bilirubin and the SLCO1B1 variant rs2900478, alkaline phosphatase and rs10740134 (REEP3), rs186021206 (ASGR2;ASGR1) and rs9987289 (LOC157273), C-reactive protein and rs2228145 (IL6R) and rs35945185 (LEPR;PDE4B), sex hormone-binding globulin (SHBG) and rs10740134 (REEP3), glycated hemoglobin (HbA1c) and rs17476364 (HK1), and triglycerides and rs1260326 (GCKR), rs112875651, rs28601761 and rs2954027 (TRIB1;LINC0086). Overall, all GRSs for the four iron status biomarkers were associated with disorders of mineral metabolism, in particular iron metabolism, as well as with red blood cell-related traits and other blood composition measures, lipid-related measurements, direct and total bilirubin, and HbA1c (Fig. 1 and Supplementary Data 17–19). Several GRSs were associated with anemias (iron, ferritin, TSP) and coagulation defects (ferritin, TSP), the GRS for TIBC was associated with liver cirrhosis without mention of alcohol, and the GRS for ferritin was associated with chronic non-alcoholic liver disease, and with phlebitis and thrombophlebitis. Finally, some of the GRSs were also associated with other continuous traits and biomarkers, including markers related to liver damage (iron, TSP, ferritin) and SHBG (iron, TSP), which is associated with liver iron overload55.
Fig. 1. Phenome-wide associations between GRSs for iron status biomarkers and 1473 phecodes, blood biomarkers, and continuous traits in the UK Biobank.
Phenome-wide associations between the GRS for each biomarker (serum iron [a], serum ferritin [b], total iron-binding capacity [c], and transferrin saturation percentage [d]) and 1473 phecodes, blood biomarkers, and continuous traits in the UK Biobank. Triangles pointing upwards indicate a positive association between the phenotype and the GRS (where a higher GRS score represents higher level of the biomarker) and vice versa. Associations with p-values < 10−324 are plotted at 10−324. The Bonferroni corrected p-value cut-off (2.3 × 10−7) is given as a red dotted line. Biological domains are indicated by color; for the significant associations, these are: hematopoietic (orange), anthropometric (dark blue), circulatory system (red), respiratory (brown), digestive (light green), genitourinary (yellow), musculoskeletal (dark green), endocrine/metabolic (magenta), symptoms (light blue). Abbreviations: MCV mean corpuscular volume, MCH mean corpuscular hemoglobin, MCHC mean corpuscular hemoglobin concentration, dist. distribution, Cong. def. congenital deficiencies, HbA1c glycated hemoglobin, SHBG sex hormone-binding globulin, LDL low-density lipoproteins, Apo B apolipoprotein B, IGF-1 insulin-like growth factor 1, FEV1 forced expiratory volume (1 s), FVC forced vital capacity, ALP alkaline phosphatase, ASAT aspartate aminotransferase, ALAT alanine aminotransferase, GGT gamma-glutamyl transferase. a: 1Other anemias, 2Impedance, 3FEV1, 4FVC, 5Total protein, 6Cystatin C, 7Phosphate, 8Apo B, 9Testosterone, 10IGF-1, 11Cholesterol, 12LDL. b: 1Anemias (iron deficiency and other anemias), 2Platelet indices (platelet count, platelet crit, platelet dist. width), 3White blood cell counts (lymphocytes, leukocytes, monocytes), 4Seated height, 5Water mass, 6Fat-free mass, 7Phlebitis and thrombophlebitis, 8Non-alcoholic liver disease, 9Gamma-glutamyl Transferase, 10Direct bilirubin, 11Urate, 12Creatinine. c: 1Mean platelet volume, 2Platelet crit, 3Platelet dist. width., 4FEV1, 5FVC, 6Liver cirrhosis, 7Cystatin C, 8Phosphate. d: 1Other anemias, 2Monocyte percentage, 3Congenital deficiency of other clotting factors, 4FEV1, 5FVC, 6ALAT, 7Cystatin C, 8Phosphate, 9LDL, 10Apo B, 11IGF-1, 12Cholesterol, 13Testosterone.
Linear Mendelian randomization
The linear MR (ratio of coefficients method) indicated an increased mortality risk with increased serum iron and TSP, with the point estimates suggesting that an increase of 1 standard deviation (SD) in both serum iron (1 SD = 6.3 µmol L−1) and TSP (1 SD = 11.3 percentage points) would lead to an increased risk of mortality of 7% (Table 3). The estimates for ferritin and TIBC were not statistically significant, however the point estimates for a 1 standard deviation increase in serum ferritin (1 SD = 46 µg L−1) and TIBC (1 SD = 9.2 µmol L−1) were a 7% increase and 4% decrease in mortality, respectively (Table 1). The estimate for ferritin was also very imprecise.
Table 3.
Linear Mendelian randomization ratio of coefficient estimates.
| Biomarker | N | Hazard ratio (95% CI) | P-value |
|---|---|---|---|
| Serum iron | 56,654 | 1.07 (1.01–1.14) | 0.03 |
| Serum ferritin* | 2335 | 1.07 (0.26–4.41) | 0.92 |
| TSP | 56,651 | 1.07 (1.02–1.12) | 0.01 |
| TIBC | 56,654 | 0.96 (0.92–1.01) | 0.10 |
Hazard ratios with 95% confidence intervals for all-cause mortality are given per 1 standard deviation increase in the biomarker. Sample size (N), Confidence interval (CI), Transferrin saturation percentage (TSP), Total iron-binding capacity (TIBC).
*Measured in fertile, non-pregnant women, 20–55 years old, with no blood donations in the two previous years.
Non-linear Mendelian randomization
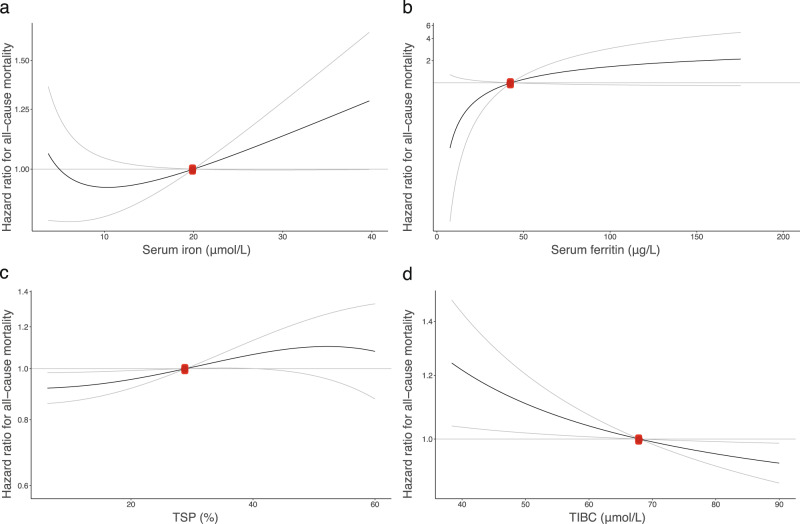
To investigate a potential non-linear causal association between iron status and all-cause mortality, we used GRSs as instruments for serum iron (F-statistic = 3,618, R2 = 0.06), TIBC (F-statistic = 8,373, R2 = 0.129), TSP (F-statistic = 6811, R2 = 0.107) and ferritin (F-statistic = 37.81, R2 = 0.015) in a non-linear MR analysis and estimated the shape of the associations between the genetically predicted traits and all-cause mortality (Fig. 2 and Supplementary Data 20–23). The median follow-up time was 23.6 years. After performing a statistical test for whether the best-fitting non-linear model of degree 1 fitted the data better than a linear model (p-values: 0.50, 0.09, 0.24, and 1 for iron, ferritin, TIBC, and TSP respectively), we generally did not find strong statistical evidence supporting a non-linear relationship over a linear one for the associations between any of the genetically proxied iron traits and all-cause mortality. However, the point estimates for serum iron did follow a J-shape, with a negative slope at very low levels of serum iron and a constant positive slope above 10 µmol L−1. The point estimates were however imprecise at the tails of the distribution. The other analyses indicated a lower risk at higher TIBC and lower TSP and ferritin levels, with a weak indication (p-value = 0.09) of a non-linear effect for ferritin. Post hoc sensitivity analyses using genetic instruments that were consistent with systemic iron status (increased iron, ferritin, and TSP, and decreased TIBC) rather than just representing a single biomarker, gave similar results (Supplementary Fig. 5).
Fig. 2. Non-linear Mendelian Randomization: causal associations between iron status biomarkers and all-cause mortality.
Dose-response curves (black) between iron traits and all-cause mortality in HUNT (gray lines give 95% confidence interval). The x-axis gives a: serum iron levels (µmol/L) [N = 56,654], b: serum ferritin (µg L−1) [N = 2335], c: transferrin saturation (%) [N = 56,651] and d: total iron-binding capacity (TIBC) (µmol L−1) [N = 56,654]. The y-axis gives the hazard ratios for all-cause mortality with respect to the reference values (red dot), which represent the established target values (iron, TIBC, TSP)84 or median value (ferritin) for the traits. The curve gradients represent the localized average causal effect at each point. N = sample size.
Discussion
We performed the largest GWAS meta-analysis to-date of iron status biomarkers and identified 123 genetic loci (78 novel for at least one biomarker) associated with iron status biomarkers, including 19 novel protein-altering variants. Although 78 loci were classified as novel for at least one of the tested biomarkers, many of them had known associations with the other tested biomarkers. Further, a high number of the loci were associated with red blood cell indices both in the current and previous studies56,57, thereby strengthening our confidence in the role of these loci in iron homeostasis. The strong associations with red blood cell indices are in line with the findings from previous studies21,39. Because iron traits are biologically linked, we might expect to find the same loci associated with several of the tested traits, as seen for loci with established roles in iron homeostasis (e.g., HFE, TF, and TMPRSS6). We confirmed the genetic similarity between iron status biomarkers by observing a high genetic correlation of 75% between serum iron and TSP, which is expected for dependent variables. However, most of the loci for TIBC and ferritin were specific for those traits, which was reflected by lower genetic correlations between some of the measured biomarkers.
Overall, our findings are consistent with established knowledge about iron homeostasis and the role of iron in various biological processes: In four of the novel loci for iron status biomarkers, the genes nearest to the index variant encoded proteins with established roles in iron regulation: (1) the mitochondrial iron transporter mitoferrin-2 (SLC25A28), (2) heme oxygenase 1 (HMOX1), which catalyzes heme degradation, (3) the iron-responsive element binding 2 (IREB2), and (4) PAS1, which regulates erythropoiesis according to cellular iron availability32. In addition to the direct associations with iron-related traits, many of the genes in both the known and novel loci were known for their role in other biological processes and diseases, such as cardiovascular or liver markers, immunity, inflammation, and cancers, suggesting that these loci might be more indirectly associated with iron status, either via processes that cause or are caused by changes in iron status. Knowing that ferritin and transferrin are also acute-phase biomarkers58, such indirect associations might also explain the high number of TIBC and ferritin-associated loci that were not associated with other iron status biomarkers. The PheWAS analyses further linked the identified loci to many different traits and phenotypes, particularly within the hematopoietic, digestive, and endocrine/metabolic domains.
The novel protein-altering variants were also found in genes associated with diverse biological traits and functions, which potentially highlight the many biological processes both involved in and dependent on iron and iron regulation. These included, but were not limited to, genes involved in or associated with: (1) Iron gut absorption, regulation, and transport (TFR2, SLC11A2, and DNAJC13)8,54,59,60, where we found a rare (minor allele frequency (MAF) = 0.0009) protein-altering variant with moderate effect size in DNAJC13, a gene suggested to be involved in transferrin recycling59. This variant was only imputed in HUNT, where it was more than 100 times more common than in other non-Finnish Europeans (https://gnomad.broadinstitute.org/variant/rs367731784)61; (2) Parkinson’s disease, which is associated with iron deposition in the brain62, and where DNAJC13 is one of several associated genes whose roles in the disease are still debated63; (3) Concentrations of hemoglobin (SPRTN, FCGR2A, CPS1, PNPLA3, and ABCA6)53,64–66, which holds more than two thirds of the body’s iron1, and bilirubin (SLCO1B1)67, which together with iron are products of heme degradation; (4) Iron-dependent (putative) tumor suppression (JMJD1C)68; (5) Fibrinolysis and bleeding (SERPINF2)69; (6) Liver-related traits (ABCA6, HNF4A, PNPLA3, and SERPINA1)69–71; and (7) Tissue specific iron accumulation (WIP1 could potentially be associated with iron accumulation in the brain via its homolog WIPI472, and the index variant we report in SERPINA1, rs28929474, has recently been proposed as a modifier of HFE-related hereditary hemochromatosis73 and as a trigger for hepatic iron overload74); A previous study also identified a different protein-altering SNP in the serine protease inhibitor SERPINA121. The transmembrane serine protease 6 (encoded by TMPRSS6) is a negative regulator of hepcidin75, a key hormone regulator of iron homeostasis17. Given the role of the transmembrane serine protease 6 in iron regulation, both serine protease inhibitors could potentially affect iron regulation via this gene. In contrast to some of the well-known genetic variants associated with iron homeostasis, most of the novel protein-altering variants in the current study were predicted as “tolerated” by in silico testing. This is however as expected for common variants, since an increased GWAS sample size will give an increased power to detect variants with smaller effect sizes than what has been identified in previous studies.
Using DEPICT, we detected an enriched expression in the liver of genes in iron status loci. This is in line with the important role of the liver in iron metabolism and storage76, including hepcidin production. Consistent with previous studies, our analysis prioritized genes encoding known hepcidin regulators such as HFE, TMPRSS6, TF, TFRC, TFR2, ERFE, and IL6R77–80. Mutations in several of these genes have been demonstrated to cause diseases of iron deficiency or overload1,19,37,81. The associations with other genes related to inflammation, both in the DEPICT and colocalization analyses, could possibly be related to the hepcidin response to inflammation. The genes and gene sets prioritized by DEPICT pointed to several different biological processes, which might reflect the numerous roles of iron in the body. A limitation with using a similarity-based method for gene set and gene prioritization for iron traits was that the software excluded the MHC region from the analysis, thereby also excluding one of the most central genes in iron homeostasis, the HFE gene.
Colocalization analysis further linked the GWAS loci both to the liver and to iron overload: iron status loci overlapped with cis-eQTLs for several of the hepcidin regulators, and genes involved in other liver functions such as lipid and fatty acid metabolism (ORMDL1, FADS1)82,83. The latter was also found in a previous independent GWAS study39 and is in line with the results from the PheWAS analysis. The colocalization of iron status loci and cis-eQTLs were however found in several tissues, and not primarily in liver. A limitation to the analysis was however due to different sample sizes for each tissue, where liver had a relatively small sample size and subsequently lower power than other tissues. Further, gene expression is highly dependent on the cellular conditions, so tissue from living donors might not produce similar outcomes.
Because iron plays an essential role in so many biological processes, several MR studies have explored the causal effect of iron status on a range of diseases22–27. Despite the known harmful effects of both iron deficiency and overload, as well as the associations of both high and low TSP with increased mortality risk in traditional observational studies13–16, no previous MR studies have investigated the shape of the exposure-outcome relationship. We, therefore, assessed the causal effect of iron status biomarkers on all-cause mortality and investigated the shape of these associations. We demonstrated that the GRSs based on the previous study were good instruments for iron, TSP, TIBC, and ferritin in the independent HUNT study (variance explained 6% (iron), 11% (TSP), 13% (TIBC), 1.6% (ferritin)), thereby validating the previous study findings. Using these, we found evidence of a harmful effect of increased serum iron and TSP (derived from serum iron and TIBC) in linear analyses that estimated population-averaged effects. The point estimates of TIBC and ferritin were also suggestive of a harmful effect of increased iron status, although the estimates were not statistically significant, and the ferritin estimate was very imprecise due to the small sample size. In non-linear models, we did not find strong statistical evidence supporting non-linear relationships over linear ones. However, there was weak evidence of a protective effect of increasing serum iron at the very low end of its distribution, at serum iron levels below the normal range of 10–34 µmol L−1 84. The results were supported by post-hoc sensitivity analyses using only genetic variants consistent with systemic iron status. Our findings confirm the previously reported associations between elevated TSP and an increased risk for overall mortality. We also found weak support for the previously reported J-shaped association between iron biomarkers and mortality, although further studies are needed to confirm this.
This study had several clear limitations. First, in our GWAS analyses we did not adjust for additional factors that could affect the biomarker concentrations, such as iron supplementation, inflammatory status, alcohol consumption, and menopausal status (except for ferritin, where the full sample was pre-menopausal). These factors could therefore have influenced the effect estimates, particularly for rare variants. Second, we would need a larger sample size to confirm a non-linear shape of the exposure-outcome relationship at the extremes of the biomarker distributions. The analysis of ferritin was particularly limited by the small sample (N = 2334) consisting of only relatively young, non-pregnant females, giving a low number of strata and (fortunately) few deaths. Third, the association of the GRSs with all-cause mortality could be attenuated because HUNT participants with suspected iron deficiency anemia or phenotypic hemochromatosis were later contacted by the primary health care services and offered treatment, and they could therefore have obtained a healthier iron status than they would otherwise have had, causing the analysis to be less precise. Finally, although the four biomarkers are commonly used to assess people’s iron status, neither of them is a very good individual predictor of iron stores, and the findings should therefore be interpreted with caution.
In summary, we have increased the number of iron status-associated loci through a large GWAS meta-analysis and validated the latest genetic risk scores for four iron status biomarkers. We find evidence of a harmful population-averaged effect of genetically proxied elevated serum iron and TSP, and weak evidence of a protective effect of increasing serum iron in individuals at the very low end of its distribution. Our findings contribute to our understanding of the genes affecting iron status and its consequences on human health.
Methods
Cohort descriptions
HUNT
The HUNT Study is a longitudinal population-based health study conducted in the county of Trøndelag, Norway since 198429. About 123,000 individuals (aged ≥ 20 years) have participated in at least one of four surveys, and more than 70,000 of these participants have been genotyped using one of three Illumina HumanCoreExome arrays: 12 v.1.0, 12 v.1.1 and 24 with custom content (UM HUNT Biobank v1.0). Samples whose genotypes had call rates < 99%, estimated contamination > 2.5%, large chromosomal copy number variants, lower call rate of a technical duplicate pair or twin, gonosomal constellations other than XX or XY, or discrepancy between inferred sex and reported gender were excluded. Following genotyping, variants with call rate < 99%, deviations from Hardy Weinberg equilibrium (p-value < 10−4 in unrelated samples of European ancestry), probe sequences that could not be perfectly mapped to the reference genome, cluster separation < 0.3, Gentrain score < 0.15, or if another assay with higher call rate had genotyped the same variant were excluded. All variants were imputed from the TOPMed reference panel (freeze 5)28 using Minimac4 v1.0 (https://genome.sph.umich.edu/wiki/Minimac4). The reference panel is composed of 53,831 multi-ethnic samples and 410,323,831 SNP and indel variants at high depth (mean read depth 38.2X). Variants with a minor allele count (MAC) > 10 or imputation R2 ≥ 0.3 were included in analysis. A subset of individuals was genotyped with additional custom content variants.
MGI
The MGI is a repository of genetic data and electronic medical records from Michigan Medicine. Approximately 80,000 participants (aged ≥18 years) have predominantly been enrolled prior to surgical procedures with over 59,000 individuals genotyped using Illumina Infinium CoreExome-24. Following genotyping, sample-level QC was performed to remove sex-mismatches, duplicates, samples with call rate < 99%, or with estimated contamination > 2.5%. Variants with GenTrain score < 0.15, Cluster Separation scores < 0.3, Hardy–Weinberg Equilibrium p-value among unrelated individuals of European ancestry <1 × 10−4, or with evidence of batch effects (p-value < 1 × 10−3, Fisher’s exact test) were excluded. Imputation was performed using the TOPMed Imputation Server (v1.2.7). Variants with MAF > 0.05% and imputation quality R2 ≥ 0.3 were included in analysis.
SardiNIA
The SardiNIA study is a longitudinal population-based health study including 6,602 individuals from the Lanusei valley on Sardinia. The participants have been genotyped on four different Illumina Infinium arrays, OmniExpress, Cardio-Metabochip85, Immunochip86, and Exome Chip). Samples with low call rate or with discrepancies between inferred and reported sex and/or relationships were excluded. After genotyping, variants with low call rates, large discordance among duplicate or identical twin genotypes, excess Mendelian inconsistencies, deviations from Hardy–Weinberg equilibrium, or MAF = 0 were excluded. Variants were then imputed from a SardiNIA-specific sequencing panel (~4× coverage) of 3839 individuals, using Minimac387. Markers with imputation quality R2 > 0.3 (or >0.6 in variants with MAF < 1%) were retained, resulting in a total of ~19 million genetic variants.
Iron status biomarkers
Distributions of the biomarker levels in the HUNT, MGI, and SardiNIA participants included in the current study are reported in Supplementary Data 2.
HUNT
Non-fasting serum samples were drawn in 1995–1997 (HUNT2). Serum iron concentration (µmol L−1) was determined using a FerroZine method using a Hitachi 911 Autoanalyzer (reagents from Boehringer, Germany). The serum transferrin concentration (µmol L−1) was analyzed by an immunoturbidimetric method using the Hitachi 911 Autoanalyzer (reagents from DAKO A/S, Denmark), and calculated for a molecular weight of 79,570 Da. TIBC was calculated as 2 × the serum transferrin concentration. The TSP was calculated as 100 × [serum iron concentration/TIBC]. Serum ferritin (µg L−1) was measured from serum samples using an Abbott AxSYM analyzer (reagents from Abbott Laboratories, USA). In total, 56,667 HUNT participants had measurements of serum iron and TIBC, 56,664 had measurements of TSP, while ferritin was only measured in 2334 women (fertile, non-pregnant, aged 20–55 years).
SardiNIA
Serum iron (µmol L−1) and serum transferrin concentrations (µmol L−1) were measured in fasting blood-samples from individuals with genotype and imputation data from the SardiNIA cohort. TIBC was calculated as 2 × the serum transferrin concentration. In total, 5930 and 5926 genotyped SardiNIA participants had measurements on serum iron and TIBC respectively.
MGI
Serum iron concentration (µmol L−1) was measured using the Ferrozine Colorimetric assay, and serum ferritin (µg L−1) was measured using a Chemiluminescent Immunoassay. Serum transferrin concentrations were measured using an Immunoturbidimetric assay, and TIBC was calculated as 2 × the serum transferrin concentration. The TSP was calculated as 100 × [serum iron concentration/TIBC]. For individuals with multiple measurements, the initial measurement was used in the analyses. In total, 10,403, 9480, 10,399, and 10,381 participants from MGI had measurements of serum iron, serum ferritin, TIBC, and TSP respectively.
Association analyses
Association analyses of all iron traits (iron, ferritin, TIBC, and TSP) in HUNT were performed using a linear mixed model regression under an additive genetic model for each variant as implemented in BOLT-LMM v2.3.488, which also controls for relatedness between the samples. Association analyses of all iron traits in MGI were performed using a linear regression model in unrelated individuals using rvtests89. In both HUNT and MGI, we applied rank-based inverse normal transformation on the iron variables after adjusting for age and sex using linear regression, and included age, sex, genotyping batch, and the first 10 principal components (PCs) of ancestry as covariates. Association analyses of serum iron and TIBC in SardiNIA were performed using age, age2, and sex-adjusted inverse-normalized residuals of TIBC or iron as input to the Efficient Mixed Model Association eXpedited (EMMAX)90 single variant test as implemented in EPACTS (https://github.com/statgen/EPACTS).
Additionally, we performed association analyses of serum iron, TIBC, and TSP in HUNT with 19,273 additional polymorphic custom content variants genotyped in 44,248 (serum iron, TIBC) or 44,246 individuals (TSP) using BOLT-LMM v2.3.488, including the same covariates and rank-based normal transformation of the variables as was done in the main analyses.
Meta-analyses
We performed fixed-effect inverse-variance weighted meta-analysis of summary statistics for iron (sample size N = 236,612), ferritin (N = 257,953), TIBC (N = 208,422) and TSP (N = 198,516) using METAL91. Serum iron and TIBC were meta-analyzed in all studies (HUNT, MGI, SardiNIA, and summary statistics from deCODE and Interval). SardiNIA did not have data on serum ferritin and TSP and was therefore excluded from these meta-analyses, while the available summary statistics for ferritin also included the DBDS study. To harmonize genomic positions from each study, we used LiftOver from UCSC92 to map the data from SardiNIA from Human Genome Build GRCh37 to GRCh38. Because standard errors were not given in the available summary statistics from deCODE, Interval and DBDS, we calculated them as the absolute value of the (effect size/qnorm(p-value/2)), where qnorm represents the inverse standard normal distribution. Prior to meta-analysis, we filtered all studies on MAF ≥ 0.001, and in HUNT, MGI and SardiNIA we performed genomic control correction of any analyses with an inflation factor λ > 1. We considered genetic loci reaching a p-value < 5 × 10−8 for follow-up analyses.
Definition of independent loci and locus novelty
Genetic loci were defined around variants with a genome-wide significant association with a trait (p-value < 5 × 10−8). The locus borders were set 500 kb to each side of the highest and lowest genetic positions reaching genome-wide significance in each region. Overlapping genetic loci were merged if the index (lowest p-value) variants were in LD (R2 ≥ 0.2 and/or D’ ≥ 0.8), or if one of the index variants was too rare to calculate LD with the other variants from our reference panel of 5000 unrelated individuals in HUNT. A locus was classified as novel for a given trait if it had not been reported previously for the trait, and novel for iron status biomarkers if it had not been previously reported for any of the four traits. Previously published variants were identified through a literature search and a look-up in the GWAS catalog93.
Annotation of genetic variants
We used plink v1.994 with a reference panel of 5000 unrelated individuals in HUNT to identify genetic variants in strong LD (R2 ≥ 0.8) with the index variants, and annotated the functional consequences and rsIDs of the index variants and LD proxies using ANNOVAR (v.2019Oct24)95 and the UCSC human genome browser92.
Functional mapping of genetic variants
We used three different bioinformatic approaches to perform functional mapping and gene prioritization of the meta-analysis summary level data: Bayesian colocalization analysis96,97, DEPICT51, and Polygenic Priority Scores98.
To assess if any iron status loci were overlapping with statistically significant (p-value < 5 × 10−8) cis-eQTL signals and consistent with shared causal variants for iron status markers and gene expression levels in specific tissue types, we used Bayesian colocalization analysis (‘coloc’) as implemented in the R package coloc. We used cis-eQTL data from 27 general tissue types (49 subtypes) in the individuals of European ancestry from the Genotype-Tissue Expression (GTEx) portal, data set v8 (https://www.gtexportal.org), and the GWAS meta-analysis results for each of the iron status biomarkers as input. For each tissue type, we analyzed all genes whose expression were associated (p-value < 5 × 10−8) with at least one iron status associated variant (p-value < 5 × 10−8), using effect sizes and standard errors for each variant-trait association as input. The coloc software estimated the variance in each trait (iron trait or gene expression level) from the sample sizes and minor allele frequencies. We set the prior probability of a genetic variant being associated with only iron traits, only gene expression, or both traits to be 10−4, 10−4, and 10−6 respectively. We considered posterior probabilities (PP4) above 75% to give support for a common causal variant for the iron trait and expression of the gene in the given tissue.
We performed gene set enrichment, gene prioritization, and tissue/cell type enrichment tests on the iron trait loci (p-value < 5 × 10−8) using DEPICT (v1.1, release 194)51. Prior to the analysis, we used LiftOver from the UCSC92 to convert the genomic positions of the genetic variants from GRCh38 to GRCh37. Enrichment results with an FDR < 5% were considered significant.
Finally, we prioritized genes by computing Polygenic Priority Scores98 from summary-level data from each iron status biomarker. The method uses Multi-marker Analysis of GenoMic Annotation (MAGMA)99 to compute gene-level associations and gene-gene correlations from the meta-analysis p-values and sample sizes and LD information from individuals of European ancestry from the 1000 Genomes reference panel100. MAGMA is applied a second time to perform enrichment analysis for genetic features. Genes are finally prioritized based on a combination of physical distance to associated genetic variants and functional similarity with other associated genes. We considered the 1% top-ranked genes per biomarker to be prioritized genes for the respective traits.
Heritability estimation
We estimated the narrow-sense additive SNP heritability of serum iron, TIBC, and TSP in HUNT using GCTA40. Ferritin heritability was not estimated because of the low sample size in HUNT. We created genetic relationship matrices (GRMs) based on 358,956 genotyped autosomal variants in 56,667 individuals with serum iron and TIBC data, and 56,664 individuals with TSP data. We used the respective GRMs with GCTA-GREML (genomic-relatedness-based restricted maximum-likelihood) to estimate the variance in each variable that was explained by the genetic variants. We used age, sex, and genotyping batch as covariates in the analyses, and we transformed the iron trait variables to normality with rank-based inverse normal transformation after regression on age and sex prior to the analyses.
Genetic correlation between iron status biomarkers
We used the LD Score Regression software41 with the iron status biomarker meta-analysis summary statistics and precomputed LD Scores for Europeans from the 1000 Genomes reference panel100, and estimated the pair-wise genetic correlations of the four iron traits. Prior to the analysis, we changed all p-values < 1 × 10−300 to the exact value 1 × 10−300 to make sure the software was able to read the smallest values and did not discard these SNPs. To ensure that only well-imputed SNPs were included in the analysis and thereby avoid bias due to variable imputation quality, we filtered the input files to the HapMap3 reference panel prior to the analysis, as recommended by the software developers (https://github.com/bulik/ldsc/).
Phenome-wide association tests (PheWAS)
We constructed GRSs for the iron status biomarkers by summing the product of the effect size and the estimated allele count (dosage) for the index variants in genome-wide significant loci (p-value < 5 × 10−8). We used TOPMed imputed estimated allele counts and effect sizes from the meta-analysis and calculated the GRS for participants of white British ancestry in the UK Biobank. We tested for pleiotropic associations of each GRS (GRS-PheWAS) and individual index variant (single variant PheWAS) with 1 394 phecodes and 79 continuous traits and blood biomarkers. We used a logistic or linear regression model respectively to assess the association of the single variant estimated allele counts (‘dosage’) or inverse normalized GRS and each phecode or continuous trait/biomarker. For the GRS-PheWAS we included as covariates sex, birth year and the first four principal components of ancestry. For the single variant PheWAS we used publicly available GWAS summary statistics (phecodes from https://pheweb.org/UKB-TOPMed/ and continuous traits and biomarkers from https://pan.ukbb.broadinstitute.org/). To correct for multiple testing, we used a Bonferroni corrected p-value significance cut-off of 2.3 × 10−7, correcting for the number of tested variants, GRSs, phecodes, biomarkers, and continuous traits. Two variants were excluded from the single variant PheWAS and 14 from the GRS-PheWAS because they were not imputed in UK Biobank (Supplementary Note 2).
Validation of genetic risk scores in HUNT
To validate the previously published results from Iceland, Denmark, and UK, we created weighted GRSs for each trait based on the published index variants and effect sizes21 using the same approach as described in the previous section. We tested the predictive ability of each GRS by regressing each trait on the respective GRS in the independent cohort, HUNT (Niron = NTIBC = 56,667, NTSP = 56,664, Nferritin = 2334), and report the trait variance explained by the GRS. In total, ten variants were excluded from the GRSs because they were not imputed in HUNT (Supplementary Note 3).
Mendelian randomization of iron status on all-cause mortality
To assess the causal association of iron status on all-cause mortality, we performed linear MR analyses using the ratio of coefficients method101, using GRSs as genetic instruments for the four iron-related traits. The GRSs were constructed as described above, using index variants and external effect sizes from the previous independent meta-analysis21. We used linear regression to estimate the associations between the iron-related traits and the GRS, and a Cox proportional hazards regression to estimate the association of the GRS with mortality. The MR estimate was obtained as the ratio of the outcome-instrument and exposure-instrument association estimates. The standard error was estimated as the standard error of the GRS-mortality association divided by the GRS-biomarker association estimate.
To further assess the shape of the association, we performed a non-linear MR with the fractional polynomial method102,103 in HUNT, using the same GRS as genetic instrument for the iron traits. The method has been described in detail elsewhere103–106. In brief, each iron-related trait was regressed on its respective GRS, including appropriate covariates, and the population was divided into 100 (iron, TIBC, TSP) or 20 (ferritin) strata based on the residual traits. We stratified on the residual traits to avoid overadjustment and collider bias, as the residual trait was defined as the residual from the regression of the biomarker on the GRS, and represented the predicted biomarker level if the GRS had been zero. The number of strata was reduced for ferritin because of the lower sample size for this biomarker. In each stratum, we used linear regression to estimate the association of the GRS with the iron trait, and Cox proportional hazards regression to estimate the association of the GRS with mortality. We calculated the localized average causal effect (LACE) of the respective trait on all-cause mortality in each stratum as the ratio of the GRS-outcome and GRS-exposure associations. Unless the fractional polynomial of degree 1 fit as good (p-value > 0.5) as that of degree 2, we plotted the best-fitting fractional polynomials of degree 2 to allow for flexibility in the non-linear biomarker-mortality relationship. Otherwise, we plotted the best-fitting fractional polynomials of degree 1. We performed meta-regression of the LACE against the mean of the exposure in each stratum and tested whether the best-fitting fractional polynomial of degree 1 fitted the LACE estimates better than a linear model using the fractional polynomial method102.
To further validate the selection of SNPs representing each biomarker in the non-linear MR, we performed post-hoc sensitivity analyses rerunning the non-linear MR method with new instruments that had stricter criteria for SNP inclusion. Here, we restricted the GRSs to index variants from the previous study21 that were not only GWAS significant for at least one trait, but also nominally significant (p-value < 0.05) for the remaining traits. Further, we excluded SNPs that had directions of effect that were not consistent with systemic iron status (increasing serum iron, ferritin, and TSP, and decreasing TIBC)107. We used the remaining 14 SNPs (Supplementary Note 4) to construct each of the four GRSs in the analysis as described for the main analysis.
Statistics and reproducibility
Unless otherwise specified, statistical analyses were performed in R v.3.6.4. Sample sizes, biomarker, age, and sex distributions per study included in the GWAS meta-analyses are given in Supplementary Data 2.
Ethics
All study participants have given informed consent. The analyses in HUNT has approval from the Norwegian Data Protection Authority and the Regional Ethics Committee for Medical and Health Research Ethics in Central Norway (REK Reference Number: 2014/144), the analyses in MGI are approved by the Institutional Review Board of the University of Michigan Medical School (IRB Reference Number: HUM00094409), the analyses in SardiNIA are approved by the local ethics committee for the Istituto di Ricerca Genetica e Biomedica-CNR (IRGB-CNR; Cagliari, Italy), and the analyses in UK Biobank are covered by the ethics approval for UK Biobank studies (application 24460) from the NHS National Research Ethics Service on 17th June 2011 (Ref 11/NW/0382) and extended on 10th May 2016 (Ref 16/NW/0274).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The Trøndelag Health Study (The HUNT Study) is a collaboration between HUNT Research Center (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health. The genotyping in HUNT was financed by the National Institutes of Health; University of Michigan; the Research Council of Norway; the Liaison Committee for Education, Research, and Innovation in Central Norway; and the Joint Research Committee between St Olav’s hospital and the Faculty of Medicine and Health Sciences, NTNU. The authors acknowledge the Michigan Genomics Initiative participants, Precision Health at the University of Michigan, the University of Michigan Medical School Central Biorepository, and the University of Michigan Advanced Genomics Core for providing data and specimen storage, management, processing, and distribution services, and the Center for Statistical Genetics in the Department of Biostatistics at the School of Public Health for genotype data curation, imputation, and management in support of the research reported in this publication. AM is funded by the EU/EFPIA Innovative Medicines Initiative Joint Undertaking BigData@Heart grant 116074. SB is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (204623/Z/16/Z). This research was supported by core funding from the: United Kingdom Research and Innovation Medical Research Council (MC_UU_00002/7), British Heart Foundation (RG/13/13/30194; RG/18/13/33946), and NIHR Cambridge Biomedical Research Center (BRC-1215-20014) [*]. *The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care.
Author contributions
M.R.M. analyzed the data and wrote the first draft of the manuscript. B.M.B., J.B.N., K.H., and C.J.W. conceived and designed the study. S.E.G., K.-H.W., A.F.H., S.A.G.T., and W.Z. contributed to the analyses. K.T. gave advice on phenotype definitions and hemochromatosis. K.H., C.J.W., F.C., D.S., and G.R.A. contributed to data acquisition. S.B., A.M., and D.G. gave advice on non-linear Mendelian randomization. M.R.M., S.E.G., K.-H.W., K.T., L.G.F., S.B., A.M., D.G., B.O.Å, B.M.B., K.H., and C.J.W. interpreted the results. All authors revised the final version of the paper.
Peer review
Peer review information
Communications Biology thanks Cassandra Spracklen, Magnus Magnusson, and Alberto Piperno for their contribution to the peer review of this work. Primary Handling Editor: Eve Rogers.
Funding
Open access funding provided by Norwegian University of Science and Technology.
Data availability
The data supporting the findings are available in the Supplementary Data and upon request. The UK Biobank data can be obtained by application (https://www.ukbiobank.ac.uk). Summary level data from previously published meta-analysis in deCODE, Interval, and DBDS are available from https://www.decode.com/summarydata/. Variant associations with phecodes, continuous traits, and biomarkers used to generate Fig. 1 are accessible from https://pheweb.org/UKB-TOPMed/ and https://pan.ukbb.broadinstitute.org/. The data underlying Fig. 2 are given in Supplementary Data 20–23. The GWAS meta-analysis summary level data from the current study are available from NTNU Open Research Data, https://dataverse.no/dataverse/ntnu (10.18710/S9TJEL).
Code availability
We used the following publicly available software and data (URLs) to generate and analyze the data: R v.3.6.4, BOLT-LMM v.2.3.4 (https://alkesgroup.broadinstitute.org/BOLT-LMM/downloads), EMMAX (https://github.com/statgen/EPACTS), METAL v.2011-03-25 (https://genome.sph.umich.edu/wiki/METAL), UCSC LiftOver command line tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver), ANNOVAR v.2019Oct24 (https://annovar.openbioinformatics.org), plink v1.9 (https://zzz.bwh.harvard.edu/plink), DEPICT (https://data.broadinstitute.org/mpg/depict), Coloc (https://chr1swallace.github.io/coloc/articles/a01_intro.html), PoPS v0.1 (https://github.com/FinucaneLab/pops), GCTA (https://cnsgenomics.com/software/gcta), LD Score Regression (https://github.com/bulic/ldsc) and SUMMnlmr (https://github.com/amymariemason/SUMnlmr).
Competing interests
The authors declare the following competing interests: D.G. is employed part-time by Novo Nordisk outside the submitted work. G.R.A. and J.B.N. work for Regeneron Pharmaceuticals. C.J.W.‘s spouse works for Regeneron Pharmaceuticals. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kristian Hveem, Cristen J. Willer, Ben M. Brumpton.
Change history
8/22/2022
Missing Open Access funding information has been added in the Funding Note.
Contributor Information
Marta R. Moksnes, Email: marta.r.moksnes@ntnu.no
Ben M. Brumpton, Email: ben.brumpton@ntnu.no
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03529-z.
References
- 1.Andrews NC. Disorders of iron metabolism. N. Engl. J. Med. 1999;341:1986–1995. doi: 10.1056/NEJM199912233412607. [DOI] [PubMed] [Google Scholar]
- 2.Rajpathak SN, et al. The role of iron in type 2 diabetes in humans. Biochim. Biophys. Acta - Gen. Subj. 2009;1790:671–681. doi: 10.1016/j.bbagen.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Belaidi AA, Bush AI. Iron neurochemistry in Alzheimer’s disease and Parkinson’s disease: targets for therapeutics. J. Neurochem. 2016;139:179–197. doi: 10.1111/jnc.13425. [DOI] [PubMed] [Google Scholar]
- 4.Jáuregui-Lobera I. Iron deficiency and cognitive functions. Neuropsychiatr. Dis. Treat. 2014;10:2087–2095. doi: 10.2147/NDT.S72491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das I, et al. Impact of iron deficiency anemia on cell-mediated and humoral immunity in children: A case control study. J. Nat. Sci. Biol. Med. 2014;5:158–163. doi: 10.4103/0976-9668.136214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahluwalia N, Sun J, Krause D, Mastro A, Handte G. Immune function is impaired in iron-deficient, homebound, older women. Am. J. Clin. Nutr. 2004;79:516–521. doi: 10.1093/ajcn/79.3.516. [DOI] [PubMed] [Google Scholar]
- 7.Silva B, Faustino P. An overview of molecular basis of iron metabolism regulation and the associated pathologies. Biochim. Biophys. Acta - Mol. Basis Dis. 2015;1852:1347–1359. doi: 10.1016/j.bbadis.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Gomme PT, McCann KB. Transferrin: Structure, function, and potential therapeutic actions. Drug Discov. Today. 2005;10:267–273. doi: 10.1016/S1359-6446(04)03333-1. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, et al. Ferritin H is a novel marker of early erythroid precursors and macrophages. Histopathology. 2013;62:931–940. doi: 10.1111/his.12101. [DOI] [PubMed] [Google Scholar]
- 10.Koury MJ, Ponka P. New insights into erythropoiesis: The roles of folate, vitamin B 12, and iron. Annu. Rev. Nutr. 2004;24:105–131. doi: 10.1146/annurev.nutr.24.012003.132306. [DOI] [PubMed] [Google Scholar]
- 11.Philpott CC. The flux of iron through ferritin in erythrocyte development. Curr. Opin. Hematol. 2018;25:183–188. doi: 10.1097/MOH.0000000000000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeiffer, C. M. & Looker, A. C. Laboratory methodologies for indicators of iron status: strengths, limitations, and analytical challenges. Am. J. Clin. Nutr.106, 1606–1614 (2017). [DOI] [PMC free article] [PubMed]
- 13.Sempos CT, Locker AC, Gillum RF, Makuc DM. Body iron stores and the risk of coronary heart disease. N. Engl. J. Med. 1994;330:1119–1124. doi: 10.1056/NEJM199404213301604. [DOI] [PubMed] [Google Scholar]
- 14.Mainous AG, III, Gill JM, Carek PJ. Elevated serum transferrin saturation, and mortality. Ann. Fam. Med. 2004;2:133–138. doi: 10.1370/afm.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stack AG, et al. Transferrin saturation ratio and risk of total and cardiovascular mortality in the general population. QJM Int. J. Med. 2014;107:623–633. doi: 10.1093/qjmed/hcu045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guedes M, et al. Serum biomarkers of iron stores are associated with increased risk of all-cause mortality and cardiovascular events in nondialysis CKD patients, with or without anemia. J. Am. Soc. Nephrol. 2021;32:2020–2030. doi: 10.1681/ASN.2020101531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lal A. Iron in health and disease: An update. Indian J. Pediatr. 2020;87:58–65. doi: 10.1007/s12098-019-03054-8. [DOI] [PubMed] [Google Scholar]
- 18.Abbaspour N, Hurrell R, Kelishadi R. Review on iron and its importance for human health. J. Res. Med. Sci. 2014;19:3–11. [PMC free article] [PubMed] [Google Scholar]
- 19.Finberg KE, et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nat. Genet. 2008;40:569–571. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benyamin B, et al. Variants in TF and HFE explain ~40% of genetic variation in serum-transferrin levels. Am. J. Hum. Genet. 2009;84:60–65. doi: 10.1016/j.ajhg.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell, S. et al. A genome-wide meta-analysis yields 46 new loci associating with biomarkers of iron homeostasis. Commun. Biol. 4, 156 (2021). [DOI] [PMC free article] [PubMed]
- 22.Gill D, et al. Associations of genetically determined iron status across the phenome: A mendelian randomization study. PLoS Med. 2019;16:1–16. doi: 10.1371/journal.pmed.1002833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gill, D. et al. Effects of genetically determined iron status on risk of venous thromboembolism and carotid atherosclerotic disease: A Mendelian randomization study. J. Am. Heart Assoc. 8, e012994 (2019). [DOI] [PMC free article] [PubMed]
- 24.Gill D, et al. The effect of iron status on risk of coronary artery disease: A Mendelian randomization study—brief report. Arterioscler. Thromb. Vasc. Biol. 2017;37:1788–1792. doi: 10.1161/ATVBAHA.117.309757. [DOI] [PubMed] [Google Scholar]
- 25.Del Greco FM, et al. Serum iron level and kidney function: A Mendelian randomization study. Nephrol. Dial. Transplant. 2017;32:273–278. doi: 10.1093/ndt/gfx164.MP143. [DOI] [PubMed] [Google Scholar]
- 26.Pichler, I. et al. Serum iron levels and the risk of Parkinson disease: A Mendelian randomization study. PLoS Med. 10, e1001462 (2013). [DOI] [PMC free article] [PubMed]
- 27.Gill D, Monori G, Tzoulaki I, Dehghan A. Iron status and risk of stroke: A Mendelian randomization study. Stroke. 2018;49:2815–2821. doi: 10.1161/STROKEAHA.118.022701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taliun D, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krokstad S, et al. Cohort profile: The HUNT study, Norway. Int. J. Epidemiol. 2013;42:968–977. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- 30.Pilia G, et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2006;2:1207–1223. doi: 10.1371/journal.pgen.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: Regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 32.Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2α expression in iron deficiency. Nat. Struct. Mol. Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- 33.Paradkar PN, Zumbrennen KB, Paw BH, Ward DM, Kaplan J. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol. 2009;29:1007–1016. doi: 10.1128/MCB.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield JB, et al. Effects of HFE C282Y and H63D polymorphisms and polygenic background on iron stores in a large community sample of twins. Am. J. Hum. Genet. 2000;66:1246–1258. doi: 10.1086/302862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Tayrac M, et al. Genome-wide association study identifies TF as a significant modifier gene of iron metabolism in HFE hemochromatosis. J. Hepatol. 2015;62:664–672. doi: 10.1016/j.jhep.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 36.Montosi G, et al. Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J. Clin. Invest. 2001;108:619–623. doi: 10.1172/JCI200113468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roetto A, et al. Hemochromatosis due to mutations in transferrin receptor 2. Blood Cells Mol. Dis. 2002;29:465–470. doi: 10.1006/bcmd.2002.0585. [DOI] [PubMed] [Google Scholar]
- 38.Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. Regulators of iron homeostasis: New players in metabolism, cell death, and disease. Trends Biochem. Sci. 2016;41:274–286. doi: 10.1016/j.tibs.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benyamin B, et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 2014;5:5926. doi: 10.1038/ncomms5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulik-Sullivan B, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poncelet L, Dumont JE, Miot F, De Deken X. The Dual Oxidase Duox2 stabilized with DuoxA2 in an enzymatic complex at the surface of the cell produces extracellular H2O2 able to induce DNA damage in an inducible cellular model. Exp. Cell Res. 2019;384:111620. doi: 10.1016/j.yexcr.2019.111620. [DOI] [PubMed] [Google Scholar]
- 43.Horton R, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 44.Gao X, et al. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J. Cell Biol. 2001;154:1161–1171. doi: 10.1083/jcb.200104058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, et al. ZNF322, a novel human C 2H 2 Krüppel-like zinc-finger protein, regulates transcriptional activation in MAPK signaling pathways. Biochem. Biophys. Res. Commun. 2004;325:1383–1392. doi: 10.1016/j.bbrc.2004.10.183. [DOI] [PubMed] [Google Scholar]
- 46.Napolitano G, Ballabio A. TFEB at a glance. J. Cell Sci. 2016;129:2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jutabha P, et al. Human sodium phosphate transporter 4 (hNPT4/SLC17A3) as a common renal secretory pathway for drugs and urate. J. Biol. Chem. 2010;285:35123–35132. doi: 10.1074/jbc.M110.121301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleming JC, et al. Characterization of a murine high-affinity thiamine transporter, Slc19a2. Mol. Genet. Metab. 2001;74:273–280. doi: 10.1006/mgme.2001.3241. [DOI] [PubMed] [Google Scholar]
- 49.Saidu Y. Physicochemical features of rhodanese: A review. Afr. J. Biotechnol. 2004;3:370–374. doi: 10.5897/AJB2004.000-2071. [DOI] [Google Scholar]
- 50.Thompson MA, et al. Human indolethylamine N-methyltransferase: cDNA cloning and expression, gene cloning, and chromosomal localization. Genomics. 1999;61:285–297. doi: 10.1006/geno.1999.5960. [DOI] [PubMed] [Google Scholar]
- 51.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu T, et al. The prognostic value of differentially expressed CYP3A subfamily members for hepatocellular carcinoma. Cancer Manag. Res. 2018;10:1713–1726. doi: 10.2147/CMAR.S159425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kichaev G, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei WQ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One. 2017;12:1–16. doi: 10.1371/journal.pone.0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gautier A, et al. Liver iron overload is associated with elevated SHBG concentration and moderate hypogonadotrophic hypogonadism in dysmetabolic men without genetic haemochromatosis. Eur. J. Endocrinol. 2011;165:339–343. doi: 10.1530/EJE-11-0215. [DOI] [PubMed] [Google Scholar]
- 56.Astle WJ, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gulhar, R., Ashraf, M. A. & Jialal, I. Physiology, Acute Phase Reactants (StatPearls Publishing, 2021). [PubMed]
- 59.Fujibayashi A, et al. Human RME-8 is involved in membrane trafficking through early endosomes. Cell Struct. Funct. 2008;33:35–50. doi: 10.1247/csf.07045. [DOI] [PubMed] [Google Scholar]
- 60.Gunshin H, et al. Slc11a2 is required for intestinal iron absorption and erythropoiesis but dispensable in placenta and liver. J. Clin. Invest. 2005;115:1258–1266. doi: 10.1172/JCI24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mochizuki H, Choong CJ, Baba K. Parkinson’s disease and iron. J. Neural Transm. 2020;127:181–187. doi: 10.1007/s00702-020-02149-3. [DOI] [PubMed] [Google Scholar]
- 63.Lunati A, Lesage S, Brice A. The genetic landscape of Parkinson’s disease. Rev. Neurol. 2018;174:628–643. doi: 10.1016/j.neurol.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 64.Vuckovic D, et al. The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182:1214–1231. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen, M. H. et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell182, 1198–1213.e14 (2020). [DOI] [PMC free article] [PubMed]
- 66.Oskarsson GR, et al. Predicted loss and gain of function mutations in ACO1 are associated with erythropoiesis. Commun. Biol. 2020;3:1–10. doi: 10.1038/s42003-020-0921-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson AD, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum. Mol. Genet. 2009;18:2700–2710. doi: 10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johansson C, et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics. 2014;6:89–120. doi: 10.2217/epi.13.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Law RHP, et al. An overview of the serpin superfamily. Genome Biol. 2006;7:1–11. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen, J. B. et al. Loss-of-function genomic variants highlight potential therapeutic targets for cardiovascular disease. Nat. Commun. 11, 6417 (2020). [DOI] [PMC free article] [PubMed]
- 71.Sookoian S, Pirola CJ. Liver enzymes, metabolomics and genome-wide association studies: From systems biology to the personalized medicine. World J. Gastroenterol. 2015;21:711–725. doi: 10.3748/wjg.v21.i3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khandia R, et al. A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: Current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells. 2019;8:674. doi: 10.3390/cells8070674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pelucchi S, et al. Hif1a: A putative modifier of hemochromatosis. Int. J. Mol. Sci. 2021;22:1–15. doi: 10.3390/ijms22031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaefer B, et al. Impaired hepcidin expression in alpha-1-antitrypsin deficiency associated with iron overload and progressive liver disease. Hum. Mol. Genet. 2015;24:6254–6263. doi: 10.1093/hmg/ddv348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang CY, Meynard D, Lin HY. The role of TMPRSS6/matriptase-2 in iron regulation and anemia. Front. Pharmacol. 2014;5:1–6. doi: 10.3389/fphar.2014.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rishi G, Subramaniam VN. The liver in regulation of iron homeostasis. Am. J. Physiol. 2017;313:G157–G165. doi: 10.1152/ajpgi.00004.2017. [DOI] [PubMed] [Google Scholar]
- 77.Darshan D, Anderson GJ. Interacting signals in the control of hepcidin expression. BioMetals. 2009;22:77–87. doi: 10.1007/s10534-008-9187-y. [DOI] [PubMed] [Google Scholar]
- 78.Arezes J, et al. Erythroferrone inhibits the induction of hepcidin by BMP6. Blood. 2018;132:1473–1477. doi: 10.1182/blood-2018-06-857995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Armitage AE, et al. Hepcidin regulation by innate immune and infectious stimuli. Blood. 2011;118:4129–4139. doi: 10.1182/blood-2011-04-351957. [DOI] [PubMed] [Google Scholar]
- 80.Lin L, et al. Iron transferrin regulates hepcidin synthesis in primary hepatocyte culture through hemojuvelin and BMP2/4. Blood. 2007;110:2182–2189. doi: 10.1182/blood-2007-04-087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pietrangelo A. Hereditary hemochromatosis—A new look at an old disease. N. Engl. J. Med. 2004;350:2383–2397. doi: 10.1056/NEJMra031573. [DOI] [PubMed] [Google Scholar]
- 82.Siow D, Sunkara M, Morris A, Wattenberg B. Regulation of de novo sphingolipid biosynthesis by the ORMDL proteins and sphingosine kinase-1. Adv. Biol. Regul. 2015;57:42–54. doi: 10.1016/j.jbior.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 83.Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human Δ-5 desaturase. J. Biol. Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- 84.Rustad P, et al. The Nordic Reference Interval Project 2000: Recommended reference intervals for 25 common biochemical properties. Scand. J. Clin. Lab. Invest. 2004;64:271–284. doi: 10.1080/00365510410006324. [DOI] [PubMed] [Google Scholar]
- 85.Voight BF, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric Traits. PLoS Genet. 2012;8:1–12. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parkes M, Cortes A, Van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 2013;14:661–673. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 87.Das S, et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Loh PR, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: An efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32:1423–1426. doi: 10.1093/bioinformatics/btw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kang HM, et al. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buniello A, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:1–16. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:1–7. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014). [DOI] [PMC free article] [PubMed]
- 97.Wallace C. Statistical testing of shared genetic control for potentially related traits. Genet. Epidemiol. 2013;37:802–813. doi: 10.1002/gepi.21765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weeks, E. M. et al. Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. medRxiv10.1101/2020.09.08.20190561 (2020). [DOI] [PMC free article] [PubMed]
- 99.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:1–19. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palmer TM, et al. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am. J. Epidemiol. 2011;173:1392–1403. doi: 10.1093/aje/kwr026. [DOI] [PubMed] [Google Scholar]
- 102.Staley JR, Burgess S. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet. Epidemiol. 2017;41:341–352. doi: 10.1002/gepi.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burgess S, Davies NM, Thompson SG. Instrumental variable analysis with a nonlinear exposure-outcome relationship. Epidemiology. 2014;25:877–885. doi: 10.1097/EDE.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burgess S, et al. Dose–response relationship between genetically proxied average blood glucose levels and incident coronary heart disease in individuals without diabetes mellitus. Diabetologia. 2021;64:845–849. doi: 10.1007/s00125-020-05377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Malik, R. et al. Relationship between blood pressure and incident cardiovascular disease: Linear and nonlinear Mendelian randomization analyses. Hypertension10.1161/HYPERTENSIONAHA.120.16534 (2021). [DOI] [PMC free article] [PubMed]
- 106.Sun YQ, et al. Body mass index and all cause mortality in HUNT and UK Biobank studies: Linear and non-linear mendelian randomisation analyses. BMJ. 2019;364:1–10. doi: 10.1136/bmj.l1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wish JB. Assessing iron status: Beyond serum ferritin and transferrin saturation. Clin. J. Am. Soc. Nephrol. 2006;1:4–8. doi: 10.2215/CJN.01490506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data supporting the findings are available in the Supplementary Data and upon request. The UK Biobank data can be obtained by application (https://www.ukbiobank.ac.uk). Summary level data from previously published meta-analysis in deCODE, Interval, and DBDS are available from https://www.decode.com/summarydata/. Variant associations with phecodes, continuous traits, and biomarkers used to generate Fig. 1 are accessible from https://pheweb.org/UKB-TOPMed/ and https://pan.ukbb.broadinstitute.org/. The data underlying Fig. 2 are given in Supplementary Data 20–23. The GWAS meta-analysis summary level data from the current study are available from NTNU Open Research Data, https://dataverse.no/dataverse/ntnu (10.18710/S9TJEL).
We used the following publicly available software and data (URLs) to generate and analyze the data: R v.3.6.4, BOLT-LMM v.2.3.4 (https://alkesgroup.broadinstitute.org/BOLT-LMM/downloads), EMMAX (https://github.com/statgen/EPACTS), METAL v.2011-03-25 (https://genome.sph.umich.edu/wiki/METAL), UCSC LiftOver command line tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver), ANNOVAR v.2019Oct24 (https://annovar.openbioinformatics.org), plink v1.9 (https://zzz.bwh.harvard.edu/plink), DEPICT (https://data.broadinstitute.org/mpg/depict), Coloc (https://chr1swallace.github.io/coloc/articles/a01_intro.html), PoPS v0.1 (https://github.com/FinucaneLab/pops), GCTA (https://cnsgenomics.com/software/gcta), LD Score Regression (https://github.com/bulic/ldsc) and SUMMnlmr (https://github.com/amymariemason/SUMnlmr).