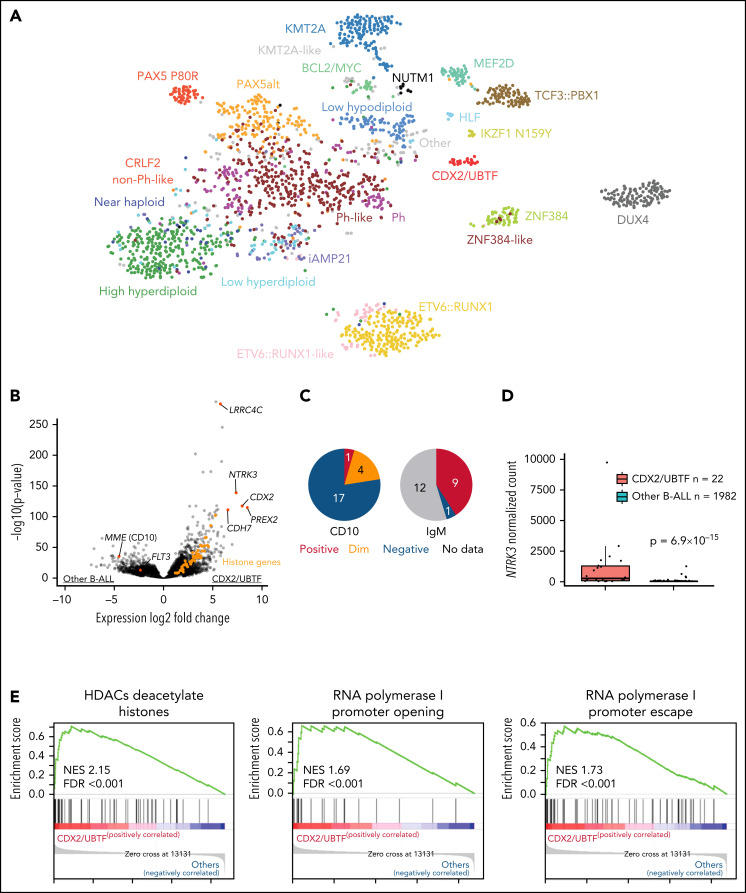
Figure 1.
Identification of a distinct subtype in B-ALL. (A) Gene expression profiling of 2004 B-ALL cases including 22 cases of CDX2/UBTF subtype (red) shown in a 2-dimensional tSNE plot. Each dot represents a sample. The top 1000 most variable genes (on the basis of median absolute deviation) were selected and processed by the tSNE algorithm with a perplexity score of 30. Major B-ALL subtypes are highlighted in different colors. (B) Differentially expressed genes in CDX2/UBTF cases compared with other B-ALL cases are shown in the volcano plot. Genes exclusively expressed in CDX2/UBTF B-ALL are colored in red and annotated. Significantly low expression of MME (CD10) and FLT3 in CDX2/UBTF B-ALL is also shown. Histone cluster genes are colored in orange showing upregulation in CDX2/UBTF B-ALL. (C) Immunophenotype of CD10 and cytoplasmic IgM in CDX2/UBTF B-ALL at diagnosis or relapse. Most cases are negative for CD10 and positive for IgM. (D) Expression of NTRK3 gene by normalized read counts comparing CDX2/UBTF and other B-ALL cases is shown. (E) Pathway analysis using top 216 differentially expressed genes (fold change >2, adjusted P < 1 × 10−30) in CDX2/UBTF B-ALL revealed enrichment of histone and ribosomal RNA related pathways.