Figure 3.
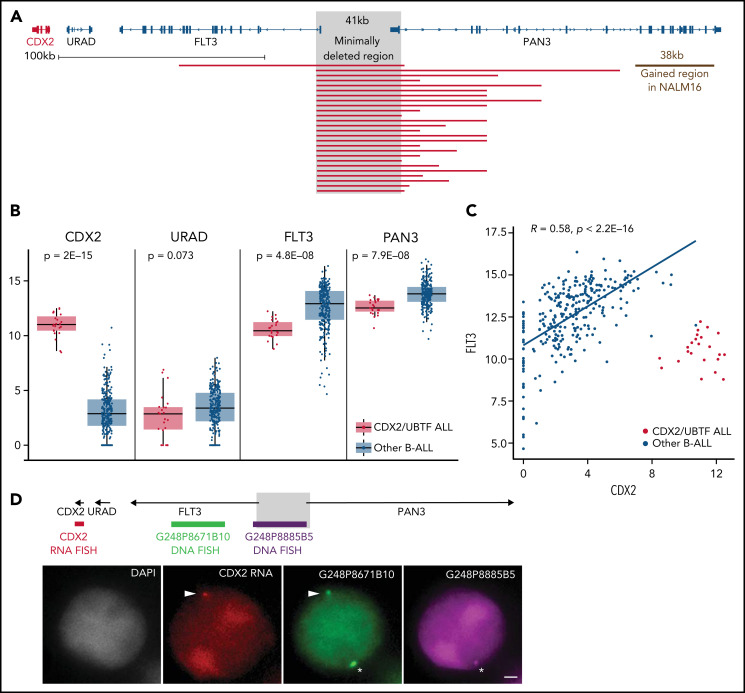
The 13q12.2 microdeletion upstream CDX2 is associated with deregulation of CDX2 in cis. (A) Capture-based sequencing of 13q12.2 locus identified focal deletion upstream of CDX2 in all CDX2/UBTF cases. The gray zone indicates the minimally deleted region. (B) Expression analysis of CDX2, URAD, FLT3, and PAN3 in CDX2/UBTF cases as compared with other B-ALL. Gene expression was quantified by RNA-seq (log2 of counts normalized using DESeq2’s median of ratios), and comparison analysis was performed by the Wilcoxon rank-sum test. (C) Dot-plot of expression data of CDX2 and FLT3 genes based on RNA-seq data for the whole cohort (n = 302) and Spearman's rank correlation analysis of non-CDX2/UBTF ALL. (D) Representative experiment (B_SL157) of sequential RNA-DNA FISH showing CDX2 primary transcripts (red signal) expressed from the allele located on the chromosome 13 exhibiting the genomic deletion (green G248P8671B10 signal and missing purple G248P8885B5 signal), shown with arrowheads. Wild-type chromosome 13 (green G248P8671B10 and purple G248P8885B5 signals) is shown by asterisks. DNA counterstained with DAPI (gray). Scale bar: 1 µm. DAPI, 4′,6-diamidino-2-phenylindole.