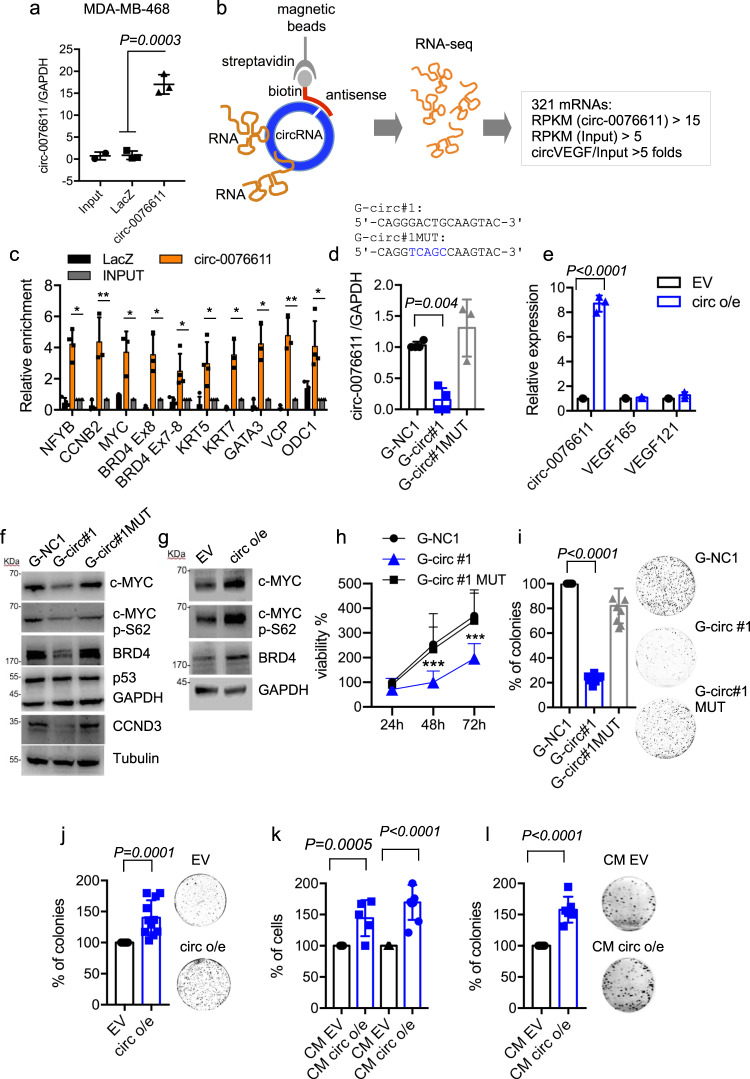
Fig. 3. Circ_0076611 impacts on cell proliferation.
a RT-qPCR analysis of circ_0076611 in ChIRP assays. Circ_0076611 has been recovered by using a biotinylated antisense oligonucleotide covering the back-splice junction, or negative control LacZ oligonucleotides. circ_0076611 expression has been normalized to GAPDH (N = 3 independent biological replicates). p value has been calculated by unpaired, two-tailed Student’s t test. b Schematic illustration of the identification of circ_0076611-interacting mRNAs by ChIRP-RNAseq. ChIRP assay was performed to recover circ_0076611 and its associated RNAs by using a biotinylated oligonucleotide complementary to circ_0076611 back- splice junction in MDA-MB-468 cells crosslinked with glutaraldehyde. Recovered RNAs were analyzed by RNAseq allowing the identification of 321 target mRNAs, considering the following settings: RPKM (circ_0076611) > 15; RPKM (input) > 5; folds circ_0076611/input > 5. c Validation of a panel of circ_0076611-mRNA interactions identified by ChIRP-RNAseq. RT-qPCR analysis of a subset of circ_0076611 target mRNAs has been carried out on RNAs recovered by ChIRP assay in MDA-MB-468 using an oligonucleotide complementary to circ_0076611 back-splice junction or control LacZ oligonucleotides. Enrichments are normalized to GAPDH mRNA and expressed as log2. *p ≤ 0.05, **p ≤ 0.005 (paired, two-tailed Student’s t test). Results from three biological replicates are shown. d Expression of circ_0076611, evaluated by RT-qPCR, in MDA-MB-468 cells transfected with circ_0076611 GapmeR oligonucleotide (G-circ#1) or control GapmeR G-circ#1 MUT, carrying a 5-nt substitution, or control GapmeR G-NC1. e Expression of circ_0076611, VEGF165 and VEGF121, evaluated by RT-qPCR, in MDA-MB-468 cells stably transfected with circ_0076611 expression vector (circ o/e) or an empty vector (EV). f, g Western blot analysis of the indicated circ_0076611 targets c-MYC, c-MYC-pSer62 and BRD4, as well as of cell cycle-related cyclin D3 (CCND3) and mutant p53 proteins in MDA-MB-468 transfected as in d, e. h Cell viability evaluated by ATPlite assay in MDA-MB-468 cell line after transfection of GapmeR oligonucleotide for circ_0076611 silencing (G-circ#1) or control GapmeR G-circ#1 MUT, carrying a 5-nt substitution, or control unrelated GapmeR G-NC1. ***p ≤ 0.0005 (paired, two-tailed Student’s t test) (N = 3). i, j Colony-formation assay of MDA-MB-468 transfected as in d, e. (paired, two-tailed Student’s t test) (N ≥ 4 independent biological replicates). k, l Cell counts (k) from MCF10A and MDA-MB-468 cells cultured with conditioned medium from circ_0076611-overexpressing MDA-MB-468 (CM circ o/e) or control cells (CM EV, empty vector) for 48 h. Colony-formation assay (l) using MDA-MB-468 cells cultured with conditioned medium from circ_0076611-overexpressing MDA-MB-468 (CM circ o/e) or control cells (CM EV, empty vector). CM was replaced every 3 days. **p ≤ 0.005 (paired, two-tailed Student’s t test) (N ≥ 4 independent biological replicates). Bars indicate standard deviation.