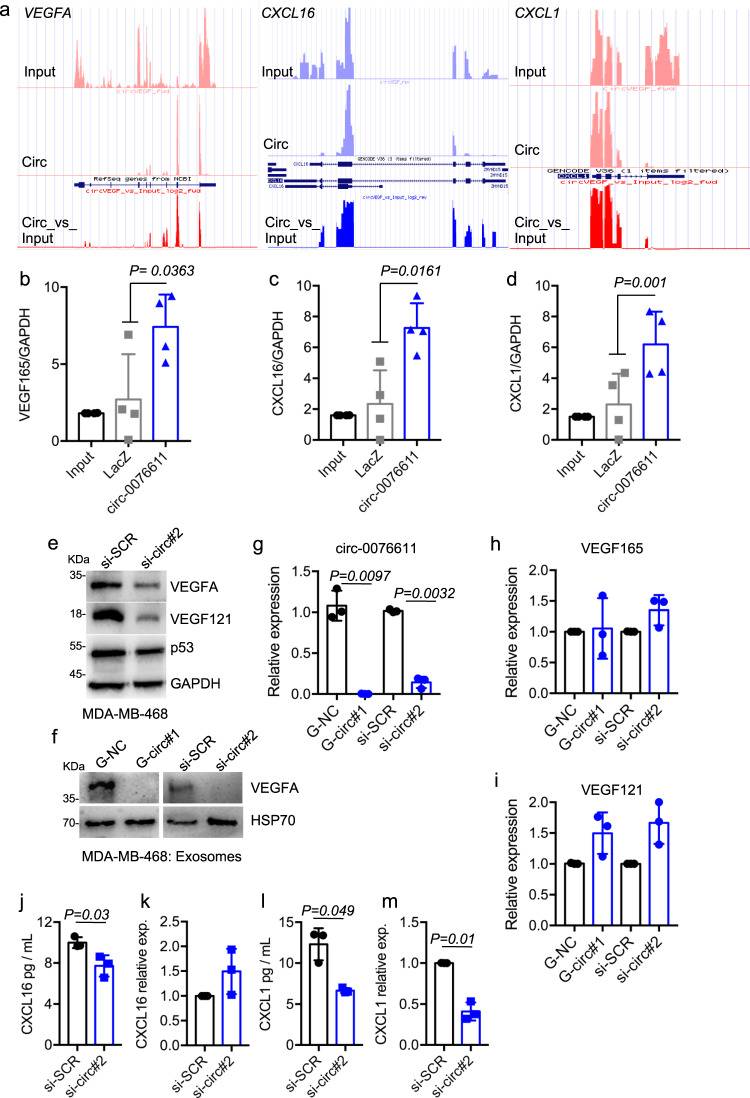
Fig. 4. Circ_0076611 controls the expression of VEGFA protein.
a Representative tracks from ChIRP-RNAseq results for VEGFA, CXCL16 and CXCL1 genes in MDA-MB-468 cells. Tracks from input and circ_0076611 (Circ) samples, along with the circ_0076611/input ratio value (Circ_vs_input) are presented. RT-qPCR analysis of VEGF165 (b), CXCL16 (c) and CXCL1 (d) mRNAs in circ_0076611 ChIRP assays performed in MDA-MB-468 cells to validate ChIRP-RNAseq results. A biotinylated oligonucleotide complementary to circ_0076611 and a LacZ oligonucleotide (as negative control) were used in ChIRP assays. Results (N ≥ 3 independent biological replicates) were normalized to GAPDH mRNA level and expressed as log2. Efficiency of circ_0076611-ChIRP is shown in Fig. 3a. (p value has been calculated by paired, two-tailed, Student’s t test). e Western blot analysis of the indicated proteins in MDA-MB-468 cells transfected with a siRNA to silence circ_0076611 (si-circ#2) or control siRNA oligo (si-SCR) for 72 h. f Western blot analysis of the indicated proteins in protein extracts from exosomes isolated from the conditioned medium of MDA-MB-468 cells transfected with GapmeRs or siRNAs to silence circ_0076611 (G-circ#1 and si-circ#2, respectively) or control oligos (G-NC, si-SCR) for 72 h. HSP70 protein has been evaluated as exosome marker. RT-qPCR analysis of circ_0076611 (g), VEGF165 (h) and VEGF121 (i) isoforms in MDA-MB-468 cells transfected with GapmeRs or siRNAs to silence circ_0076611 (G-circ#1 and si-circ#2, respectively) or control oligos (G-NC, si-SCR) for 72 h. p value has been calculated by paired, two-tailed, Student’s t test on N = 3 independent biological replicates. Expression of CXCL16 and CXCL1 proteins and mRNAs, evaluated, respectively, by ELISA assay (j–l) and RT-qPCR (k–m), in MDA-MB-468 cells transfected with a siRNA to silence circ_0076611 (si-circ#2) or control siRNA oligo (si-SCR) for 72 h. ELISA assay has been performed on conditioned media from MDA-MB-468 cells. p value has been calculated by paired, two-tailed, Student’s t test on N = 3 independent biological replicates. Bars indicate standard deviation.