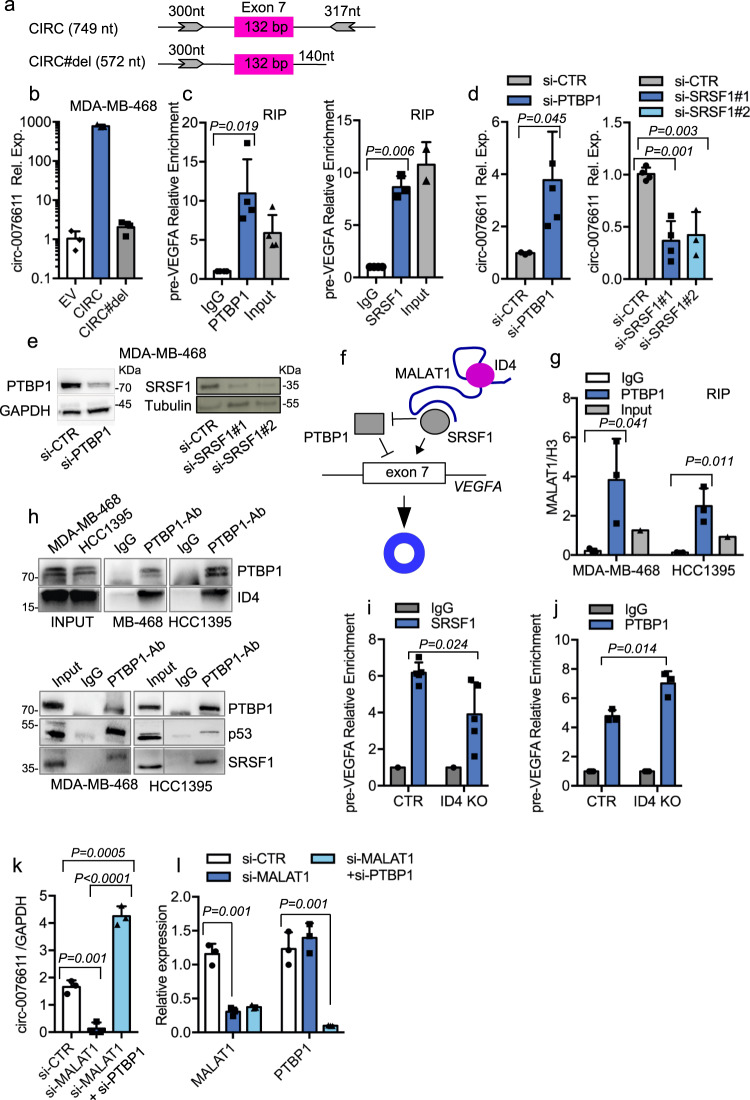
Fig. 7. MALAT1 and ID4 favor circ_0076611 expression by counteracting RNA-binding protein PTBP1.
a Schematic representation of the genomic regions cloned in pCDNA3.1 vector to generate a circ_0076611 expression vector (indicated as CIRC) and a deletion mutant lacking an inverted repeated sequence required for circ_0076611 expression (indicated as CIRC#del). b Expression of circ_0076611 (log10), evaluated by RT-qPCR, in MDA-MB-468 cells transiently transfected with a pCDNA3.1 vector (EV) or with the circ_0076611 expression vectors indicated in a. c Ribonucleoprotein Immunoprecipitation assays (RIP) in MDA-MB-468 cells (after UV crosslinking) showing the enrichment of VEGFA precursor RNA (pre-VEGFA) in samples immunoprecipitated with control IgG or with antibodies directed to PTBP1 (left) or SRSF1 (right) proteins. Data are normalized to 28S rRNA and presented as folds of enrichment over IgG. d, e Expression of circ_0076611, evaluated by RT-qPCR, in MDA-MB-468 cells transfected with a pool of siRNAs directed to PTBP1 (left) or two different siRNAs directed to SRSF1 for 72 h (right) (d). WB showing the efficiency of PTBP1 and SRSF1 interference after 72 h of siRNA transfection is shown in e. f Scheme summarizing the regulatory roles of lncRNA MALAT1 and of proteins ID4, SRSF1 and PTBP1 in the control of circ_0076611 back-splicing. g, h RIP assay in the indicated cell lines (after UV crosslinking) showing the level of MALAT1 lncRNA, evaluated by RT-qPCR, in samples immunoprecipitated with control IgG or with an antibody directed to PTBP1 protein (g). A fraction (25%) of eluates recovered during RIP experiments has been used for protein analysis by WB to control PTBP1 IP and co-IP of ID4, mutant p53 and SRSF1 proteins (h). Enrichment of SRSF1 (i) and PTBP1 (j) proteins on pre-VEGFA mRNA evaluated by RIP assay in control MDA-MB-468 cells (CTR) and in ID4 knocked-out counterpart (ID4-KO) (after formaldehyde crosslinking) by using antibodies directed to SRSF1 or PTBP1 and IgG as negative control. Data are presented as folds of enrichment over IgG. Expression of circ_0076611 (k) and MALAT1 (l) and PTBP1 (l), evaluated by RT-qPCR, in MDA-MB-468 cells transfected with siRNAs directed to MALAT1, or to MALAT1 and PTBP1, for 72 h. Data are presented as mean plus standard deviation. p values have been calculated by paired, two-tailed Student’s t test. Results from at least three biological replicates are shown. Bars indicate standard deviation.