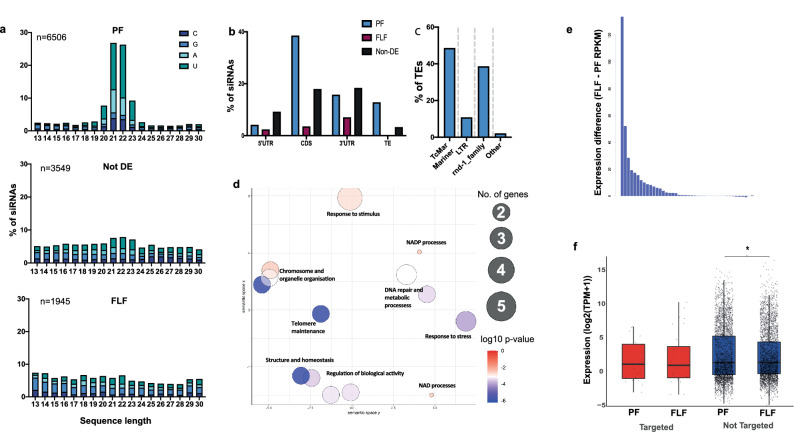
Figure 2.
21–22Us with a 5′ monophosphate associated with parasitism. (a) Length distribution and 5′ starting nucleotide of 13–30 nt sRNAs with a 5' modification originating from either protein-coding genes, intergenic spaces or transposable elements, that are significantly more abundant in the PF (n = 6506 sequences), FLF (n = 1945 sequences) or not DE (n = 3549 sequences). (b) Predicted targets of significantly more abundant 21–22Us in the PF, FLF and non-DE based on antisense sequence complementarity. Targets include protein-coding genes (CDS, 5′UTR and 3′UTR) and TE sequences. (c) Classification of predicted TE targets of 21–22Us in the PF. Targets include TcMar-Mariner DNA transposon (48.5%), LTR retrotransposons (10.8%) and an unclassified family (38.6%). (d) Enriched GO terms based on the biological function (BF) of the 21–22U predicted target genes. Clustering of the GO terms based on similar functions was carried out using REVIGO. Size of circle indicates the number of genes and colour indicates p-value. (e) Difference in expression level (RPKM: FLF minus PF) of genes putatively targeted by PF-overexpressed 21–22Us. The targeted genes were more highly expressed in the FLF cf. PF stage. (f) Expression of TEs either predicted to be targeted or not targeted by PF-overexpressed 21-22Us. *Indicates p < 0.01. RPKM = Reads per kilobase million; TPM = Transcripts per kilobase million.