Extended Data Fig. 3.
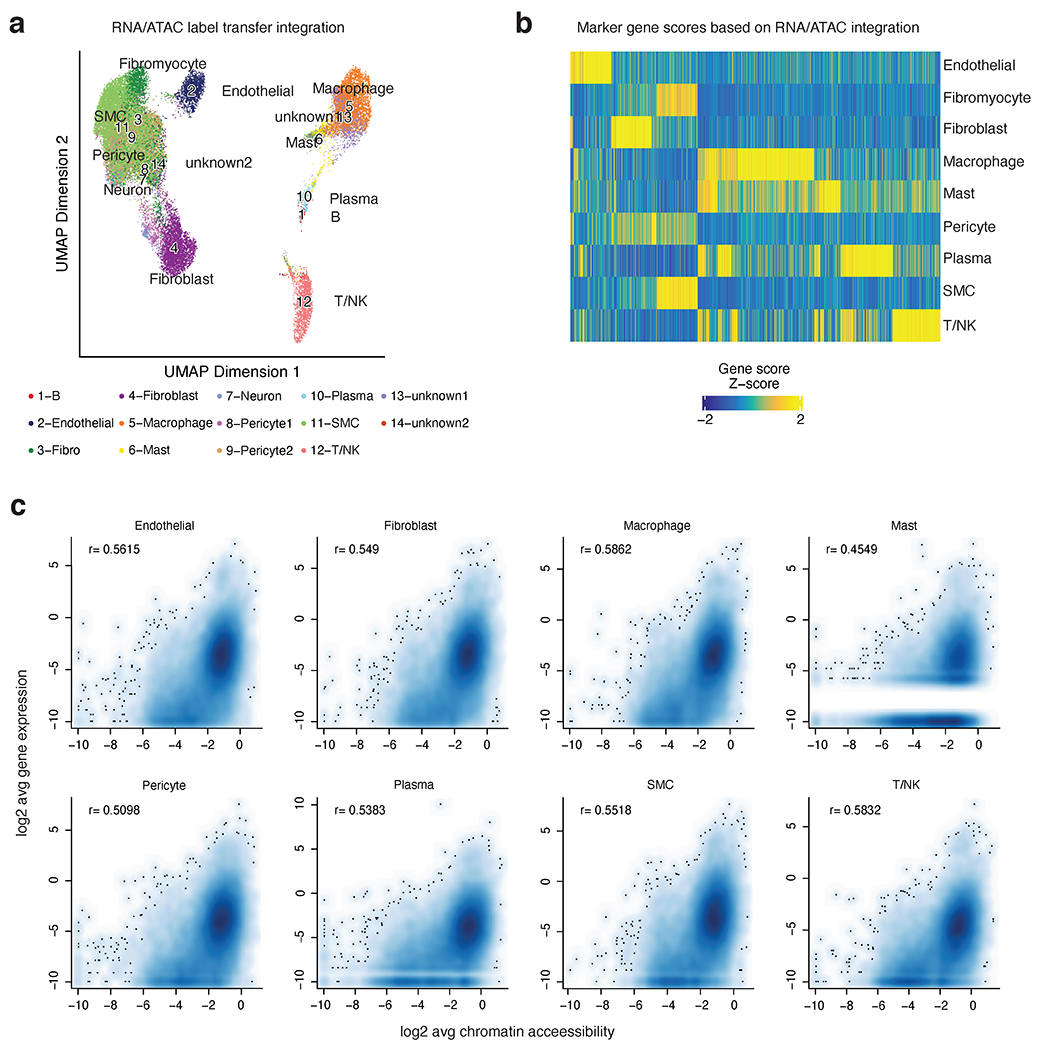
Integration of human coronary artery snATAC-seq data with human coronary artery scRNA-seq (from Wirka et al. Nat Med 2019). (a) UMAP showing projection of scRNA-seq cluster labels onto cells in the snATAC-seq dataset. Colors represent the assigned cellular identities from scRNA-seq label transfer. Detailed parameters of the snATAC-seq/scRNA-seq integration are provided in the Methods section. (b) Heatmap of marker gene scores after ArchR RNA/ATAC integration highlights 4,649 marker features. (c) Correlation of cell type specific scRNA and snATAC promoter accessibility (pseudo bulk reads from ATAC signal centered on TSS (+/− 3kb) for each gene). Log2 transformed data is represented as scatter plots and Pearson correlation coefficients are shown for each cell type. White lines represent missing gene counts from scRNA-seq dataset, which is most apparent in the low abundant Mast cells.
