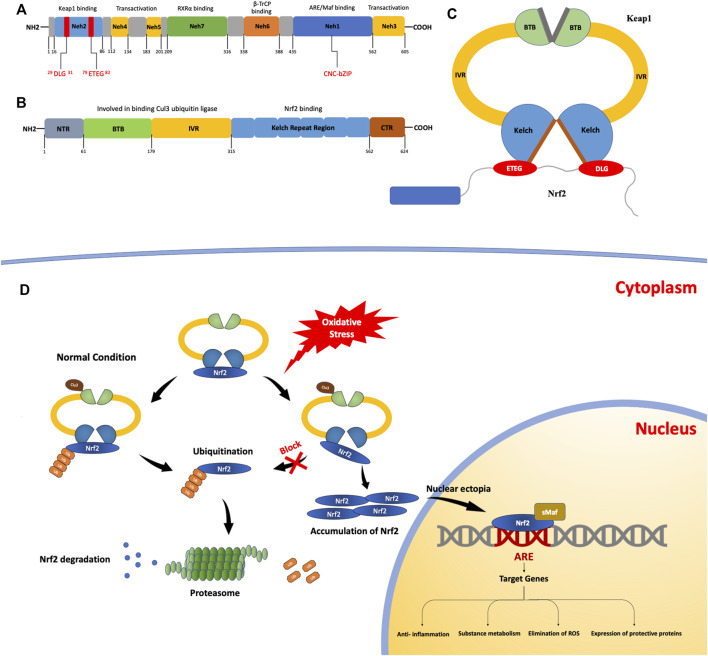
FIGURE 1.
The structures and processes involved in fibrosis. (A) Structure of the nuclear erythroid 2-related factor 2 (NRF2) protein. The NRF2-epichlorohydrin (ECH) homology (Neh) 1 domain contains a base sequence of cap-n-collar type basic leucine zipper DNA, which assists in the nuclear transfer of NRF2. The Neh2 domain contains two motifs that bind to Kelch-like ECH-associated protein 1 (KEAP1): ETGE motifs with high affinity and DLG motifs with low affinity. (B) The structure of the KEAP1 protein. KEAP1 is a homodimer. (C) It contains a Kelch repeat region that combines two motifs (ETGE and DLG) with different affinities to form a hinge–latch structure. The ETGE motif has high affinity and is considered a “hinge”. The DLG motif has a low affinity and can be regarded as a “latch”. (D) The KEAP1/NRF2 signaling pathway. Under normal conditions, NRF2 binds to KEAP1 to form an ɑ-helical conformation containing seven lysine residues and then acts as a target for the ubiquitination and cleavage of NRF2. After the ubiquitination of NRF2 by Cullin3 (Cul3), it is degraded and free KEAP1 is involved in the next round of NRF2 binding and ubiquitination. When the oxidation equilibrium is disrupted, the binding of NRF2 and KEAP1 is also affected. The “lock” structure, namely the binding between the DLG motif and the Kelch repeat domain, is destroyed. Therefore, NRF2 ubiquitination is blocked and NRF2 degradation is insufficient. As NRF2 continues to accumulate, free NRF2 is translocated to the nucleus. It can bind with antioxidant response elements to induce the expression of target genes, thus activating the body’s antioxidant defense and anti-inflammatory systems.