Fig. 1. NFIA and NFIX cooperatively silence HBG1/2 genes in HUDEP2 cells.
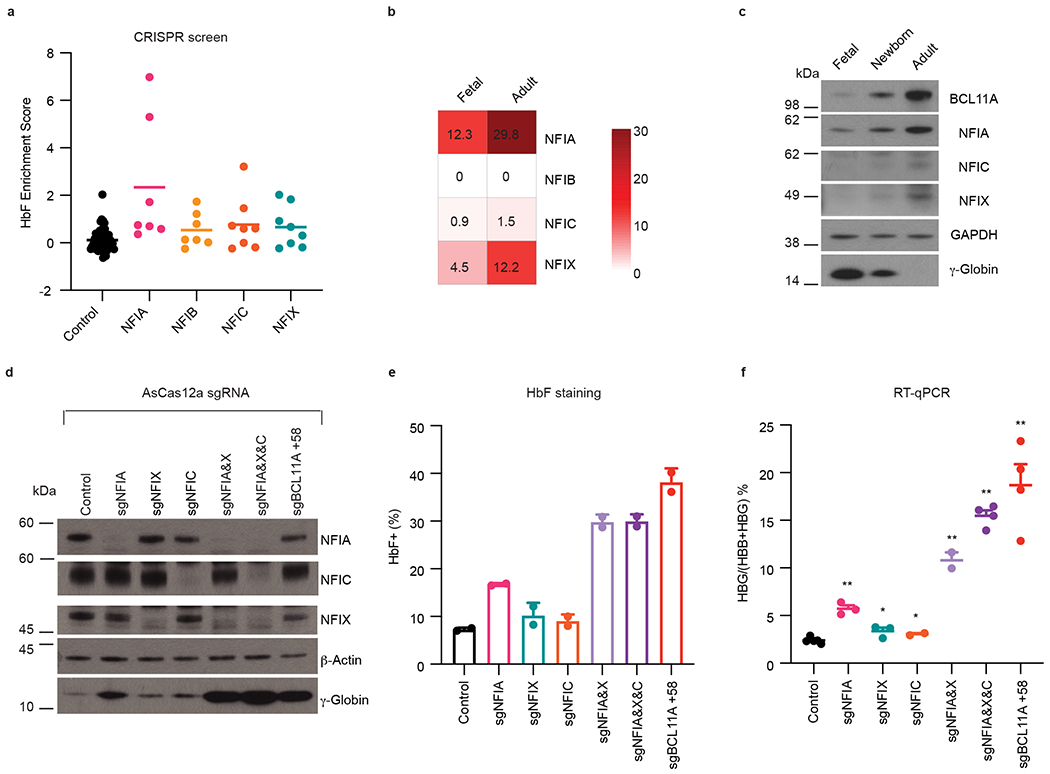
a, Enrichment scores of negative control sgRNAs (n = 12) and sgRNAs targeting NFI factors (NFIA, n = 7; NFIB, n = 7; NFIC, n = 8; NFIX, n = 8) in DNA binding domain-focused CRISPR-Cas9 screen. Data were obtained from Huang et al9. n represents different sgRNAs. b, Heatmap of NFI gene expression levels in primary erythroblasts from fetal liver (fetal, expressing HBG, n = 12) or bone marrow (adult, expressing HBB, n = 12). Fragments per kilobase of transcript per million mapped reads (FPKM) of indicated genes were obtained from Lessard et al.24. n represents different donors/biological replicates. c, Representative immunoblots of NFI proteins in erythroblasts derived from fetal liver (fetal, expressing HBG), cord blood (newborn, expressing both HBG and HBB), and peripheral blood (adult, expressing HBB). BCL11A and γ-globin served as positive controls and GAPDH as loading control. Experiments were performed twice with similar results. d-f, HUDEP2 cells expressing CRISPR-AsCas12a were transduced by a lentiviral vector carrying control sgRNA or sgRNAs targeting indicated NFI genes or the BCL11A +58 enhancer. Indicated HUDEP2 cells were analyzed at the end of differentiation (day 5). d, Representative immunoblots of NFI proteins and γ-globin in indicated HUDEP2 cells. β-Actin served as loading control. Experiments were performed three times with similar results. e, HbF+ fraction (%) of control (n = 2) and indicated gene edited HUDEP2 cells (n = 2) as determined by intracellular HbF staining. n represents two biological replicates. Data are expressed as mean ± SEM. f, The ratio of HBG/(HBG+HBB) of indicated HUDEP2 cells as determined by RT-qPCR (control, n = 5; sgNFIA, n = 3; sgNFIX, n = 3; sgNFIC, n = 2; sgNFIA&X, n = 2; sgNFIA&X&C, n = 4; sgBCL11A +58, n = 4). Data are expressed as means ± SEM. n represents biological replicates. * P < 0.05, ** P < 0.01. P values were calculated by comparing indicated samples to control with parametric unpaired two-tailed Student’s t-test. sgNFIA, P < 0.0001; sgNFIX, P = 0.0248; sgNFIC, P = 0.0426; sgNFIA&X, P < 0.0001; sgNFIA&X&C, P < 0.0001; sgBCL11A +58, P < 0.0001.
