Extended Data Fig. 3.
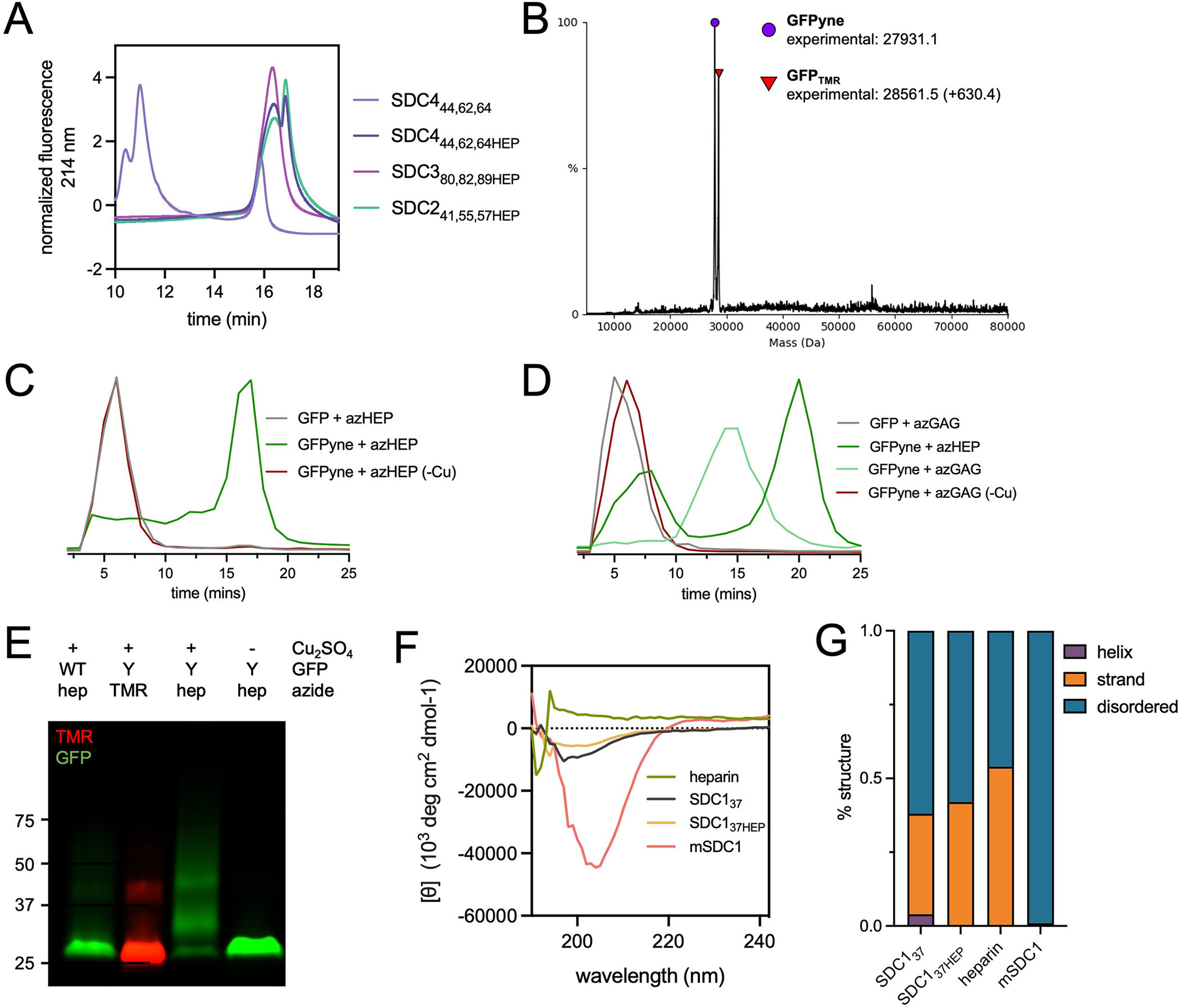
Glycoconjugation of pPY-modified proteins by CuAAC (A) Weak anion exchange (WAX) traces demonstrating successful conjugation of azHEP to form SDC241,55,57HEP (mint), SDC380,82,89HEP (pink) and SDC444,62,64HEP (dark purple). SDC444,62,64 core protein (light purple) included as reference point for SDC core proteins (. (B) Intact mass spectrometry of pPY-modified GFP (GFPyne(His)6), used as a model protein, confirms the appropriate mass shift (+630.4 Da) when reacted with tetramethylrhodamine (TMR)-azide. (C) WAX traces after reaction of GFPyne(His)6 with azHEP demonstrates a shift to the more anionic product (dark green). When the reaction is performed without copper (-Cu, red), the trace overlaps with WT GFP (grey). (D) WAX traces after reaction with azGAG (green) demonstrate a less anionic glycoconjugate than GFPyne + azHEP (dark green). Copper-free controls overlap with WT GFP peak. (E) SDS-PAGE analysis of WT and GFPyne(His)6 (Y) confirms the incorporation of pPY by TMR detection only in Y. azHEP conjugation results in a higher molecular weight smear, only in the presence of copper. Representative of two technical replicates, molecular weight ladder indicates kDa. (F) Circular dichroism spectra of heparin (red), SDC137 (grey), SDC137HEP (orange) and HEK239T expressed mSDC1 (pink). (G) Analysis of the predicted structures from CD spectra indicates that glycosylation of SDC137 does not impart a change, or increase, in protein structures and native, glycosylated SDC1 is highly disordered.
