Figure 1.
Study design, genetic distance analysis, and distribution of genomic variants across the genome
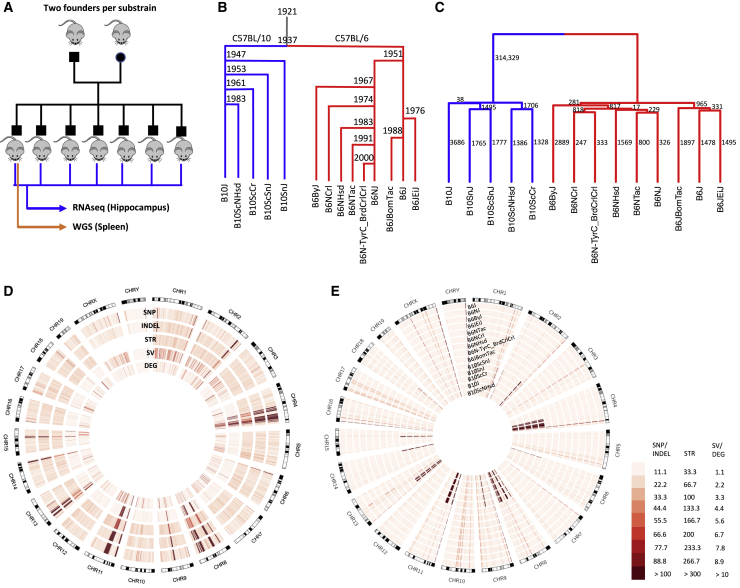
(A) The design of our study. Mice from nine C57BL/6 and five C57BL/10 substrains were purchased from four vendors. For each substrain, between six and eleven male offspring from the first generation born in our colony were chosen for hippocampal RNA sequencing (RNA-seq). One male offspring per substrain was chosen for whole-genome sequencing (WGS) using DNA extracted from the spleen.
(B) The historical relationship of C57BL/6 and C57BL/10 substrains is illustrated as a tree (Charles River Labs at https://www.criver.com/; Jackson Laboratory at https://www.jax.org/).37
(C) Dendrogram showing the similarity of the substrains based on genomic variants, including SNPs (LD-pruned, indels not included), STRs, and SVs. The numbers beside each branch indicate the number of SNPs separating the substrains in the subtree below the branch from all the other substrains.
(D) Circos plot showing the SNPs, indels, STRs, SVs, and DE genes across the genome for 14 C57BL/6 and C57BL/10 substrains. Regions with a high density of polymorphisms (hot spots) on chromosomes 4, 8, 9, 11, and 13 are obvious (see also Figures S2 and S3).
(E) Circos plot showing SNPs with non-reference genotypes for each substrain. This plot shows that most hot spots in (D) are due to regions where all C57BL/6 differ from all C57BL/10 substrains (see also Figures S2 and S3). A few regions where all substrains (including C57BL/6J) do not match the reference are also evident.