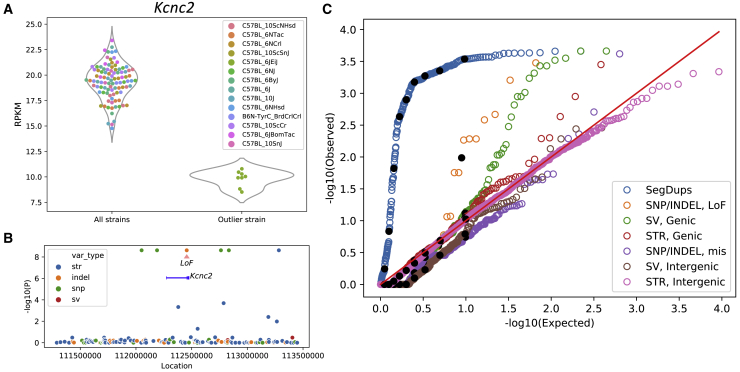
Figure 2.
Association of gene expressions with genomic variants
(A) Expression of Kcnc2 is lower for C57BL/6JEiJ compared with the other substrains.
(B) cis-variants of Kcnc2 are tested for association with the median expression by the linear regression model. One indel is a frameshift loss-of-function variant and therefore has a strong prior to be the causal variant. In addition to that, four SNPs and one STR also have the same strain distribution pattern and therefore the same –log10 (p) value. For most of the DE genes, no variant belonged to a class that had a strong prior for causality. In those cases, any of the variants with the smallest p values (or a combination thereof, or more distant variants) might be causal (see also Figure S4).
(C) The distribution of the p values of variants in different categories is compared against the uniform distribution in a QQ plot.
A linear mixed model is used with the genomic relatedness matrix (GRM) as a random effect to control for population structure, and the parental strain (C57BL/6 or C57BL/10) is used as a fixed effect to identify associations within C57BL/6 and C57BL/10 substrains. The black dots show the deciles of the data in each category. The SegDup category includes associations between the copy number variation of the DE genes intersecting with SegDup regions (obtained by read depth across the segmental duplication regions of the reference genome) and the gene expression. Loss-of-function and missense mutations are two categories of SNP/indels. Genic SVs include those intersecting with gene features such as exons, TSSs, UTRs, promoters, enhancers, and introns, and genic STRs include those intersecting with exons, TSSs, 5’ UTRs, and promoters. Intergenic SVs and STRs are those not intersecting with any gene features and are paired with a gene with the closest TSS (see also Table S2).