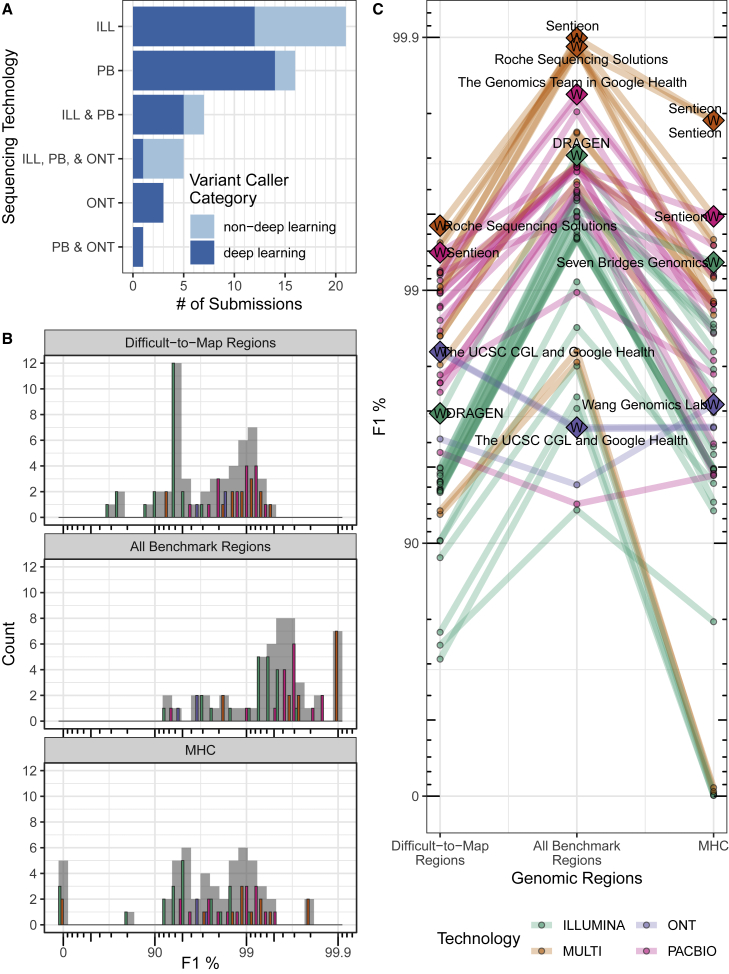
Figure 2.
Challenge submission breakdown and performance overview
(A) Challenge submission breakdown by technology and type of variant caller used. Deep-learning methods use either a convolutional neural network or a recurrent neural network architecture for learning the variant-calling task, while non-deep-learning methods use techniques that broadly arise from statistical techniques (e.g., Bayesian and Gaussian mixture models) or other ML techniques (e.g., random forest) to differentiate variant and non-variant loci based on expert-designed features of the sequencing data.
(B and C) Overall performance (B) and submission rank (C) varied by technology and stratification (log scale). Generally, submissions that used multiple technologies (MULTI) outperformed single-technology submissions for all three genomic context categories. (B) A histogram of F1 percentage (higher is better) for the three genomic stratifications evaluated. Submission counts across technologies are indicated by light gray bars and individual technologies by colored bars. (C) Individual submission performance. Data points represent submission performance for the three stratifications (difficult-to-map regions, all benchmark regions, MHC), and lines connect submissions. Category top performers are indicated by diamonds with Ws and labeled with team names. F1 is plotted on a phred scale with axes labels and ticks indicating F1 percentage values.