Abstract
Rabies infection is nearly 100% lethal if untreated and kills more than 50,000 people annually, many of them children. Existing rabies vaccines target the rabies virus glycoprotein (RABV-G) but generate short-lived immune responses, likely because the protein is heterogeneous under physiological conditions. Here, we report the 3.39 Å cryo–electron microscopy structure of trimeric, prefusion RABV-G complexed with RVA122, a potently neutralizing human antibody. RVA122 binds to a quaternary epitope at the top of RABV-G, bridging domains and stabilizing RABV-G protomers in a prefusion state. RABV-G trimerization involves side-to-side interactions between the central α helix and adjacent loops, rather than contacts between central helices, and interactions among the fusion loops at the glycoprotein base. These results provide a basis from which to develop improved rabies vaccines based on RABV-G stabilized in the prefusion conformation.
Fusion loops and neutralizing antibody RVA122 stabilize rabies virus glycoprotein in its prefusion, trimeric conformation.
INTRODUCTION
Untreated rabies infections are nearly 100% fatal, causing 50,000 to 60,000 human deaths annually and significantly affecting animal populations (1). Rabies vaccines for human use consist of inactivated virus and have existed since the late 1800s but do not elicit lifelong immunity despite being fully protective in the short term (6 months to a year after vaccination). In humans, levels of vaccine-elicited neutralizing antibodies usually wane 1 to 5 years after vaccination (2), and frequent revaccination is required to maintain neutralizing antibody titers understood to provide protection against rabies infection (3). Unvaccinated individuals who are exposed to rabies are given postexposure prophylaxis consisting of polyclonal antibodies derived from sera of vaccinated individuals or immunized horses and multiple doses of the rabies vaccine (4). However, both frequent vaccination and postexposure treatment are unaffordable in low-income countries where most rabies deaths occur.
Rabies virus is a member of the family Rhabdoviridae and the genus Lyssavirus. Lyssaviruses are further divided into three phylogroups, with phylogroup I comprising 12 different species including the rabies virus prototype species and phylogroups II and III containing more distantly related species (5). Most phylogroup I lyssaviruses are believed to cause viral encephalitis in humans and the same clinical symptoms as rabies infection. The rabies virus glycoprotein (RABV-G) is the only surface-exposed protein on the virus and is the target of vaccine-elicited neutralizing antibodies. RABV-G shares 57 to 78% protein sequence identity with other lyssaviruses but only ~20% identity with other rhabdoviruses. Pan-lyssavirus antibodies that recognize conserved glycoprotein epitopes are desirable for developing more potent and broadly effective therapeutics, but thus far, only a small number have been described in detail (6–9).
On the viral surface, RABV-G is structurally heterogeneous and only a portion is recognizably trimeric (10–12). RABV-G undergoes reversible, largely pH-dependent transitions between pre- and postfusion conformations (10, 13, 14). In the prefusion conformation, the fusion loops point toward the viral membrane (7, 12). Exposure to acidic pH triggers a conformational change to the postfusion state during which RABV-G elongates and the fusion loops point away from the viral membrane and toward the target cell membrane (7, 12). Virions display both pre- and postfusion conformations over a range of physiological pHs due to the reversible, pH-dependent, conformational equilibrium of the glycoprotein (13, 15, 16). This structural heterogeneity may affect the generation of neutralizing antibodies that often target quaternary epitopes and may contribute to the poor longevity of the postvaccination immune response.
Here, we report the structure of trimeric, wild-type RABV-G bound to the human antibody RVA122, which was isolated from a vaccinated individual as part of an effort to develop neutralizing, pan-lyssavirus antibodies for improved postexposure treatment (8). RVA122 is specific for the RABV-G prefusion conformation and potently neutralizes multiple phylogroup I lyssaviruses, including rabies (8). We show that the RABV-G trimeric interface involves interactions between the central α helix and adjacent loops and demonstrate the role of the fusion loops in trimerization and stabilization of the prefusion conformation. The prefusion trimer structure, elucidation of a potently neutralizing antibody epitope, and illumination of the fusion loop structure and activity detailed here can be used to guide the design of improved vaccines and to identify therapeutic drug targets.
RESULTS
Structure determination
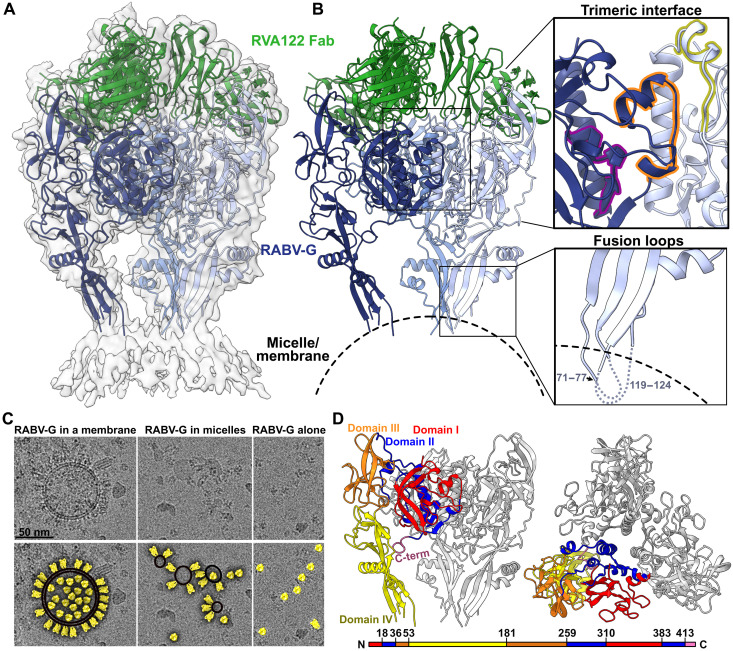
We used cryo–electron microscopy (cryo-EM) to resolve the structure of the trimeric RABV-G ectodomain (PV strain) in complex with three RVA122 antigen-binding fragments (Fabs) to 3.39 Å (Fig. 1, fig. S1, and table S1). RABV-G fusion loops anchor to either cellular membranes that copurify with the protein or detergent micelles added during grid preparation (Fig. 1, A to C). RABV-G trimers with unanchored fusion loops were also present on grids (Fig. 1C), but the fusion loops were too flexible to reach high resolution. Anchored fusion loops also adopted multiple conformations (movie S1).
Fig. 1. Structure of the prefusion RABV-G trimer bound to neutralizing antibody RVA122.
(A) Molecular model of RABV-G (blue) and RVA122 Fab (green) complex fitted into the corresponding cryo-EM density map. (B) RABV-G trimer with magnified views of the trimeric interface (top right) and fusion loops (bottom right). In the trimeric interface, the small helix (purple), bracket loop (orange), and corkscrew loop (yellow) are highlighted. (C) Top: Raw micrographs of glycoprotein complexes embedded in cellular membranes (left), embedded in micelles (center), or unembedded (right). Bottom: RABV-G (yellow), membranes (black, double outline), and micelles (black, single outline) are indicated. (D) Side view (left) and top view (right) of the RABV-G trimer, with domains I to IV and the C terminus indicated for one protomer. Colors correspond to RABV-G domains I to IV, also shown in the schematic diagram of the RABV-G sequence.
RABV-G protomers have four linked domains (Fig. 1D) that reposition via hinge regions during the prefusion to postfusion transition (7). Domains I and III comprise the solvent-exposed upper half of RABV-G and contain multiple antigenic sites (including the RVA122 epitope) and receptor binding sites (Fig. 1D and fig. S2) (8, 17–19). Domain II contains a central helix that elongates in the postfusion transition (7), and domain IV contains the two fusion loops. Domains I and II are also known as the central domain, domain III as the Pleckstrin homology domain, and domain IV as the fusion domain.
Trimeric interface
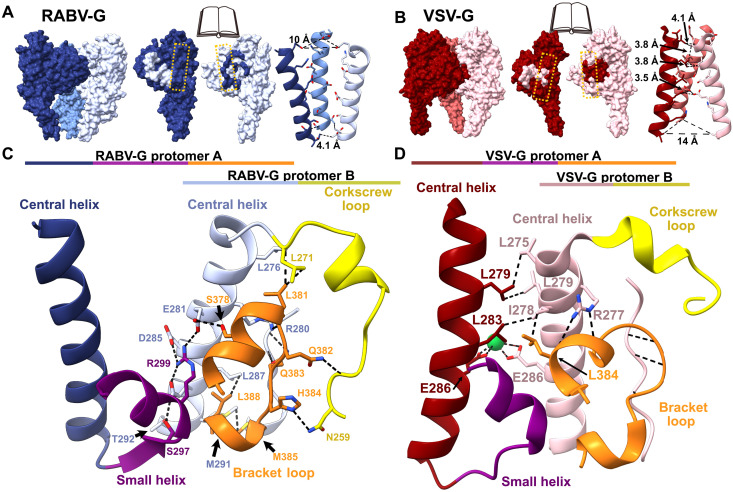
Although an α helix from each RABV-G protomer forms a threefold axis of symmetry at the center of the glycoprotein (Fig. 1D), the helices are too distant to interact (Fig. 2A). Instead, the RABV-G trimeric interface primarily consists of lateral interactions among protomers (Fig. 2, A and C). Key to these interactions is a loop in domain I that forms a bracket between two short helices (residues 378 to 384). This bracket loop extends out from one protomer to interact with two sections of the neighboring protomer: The central helix (residues 274 to 293) and a loop that extends from the central helix to form a corkscrew (residues 259 to 271; Fig. 2C). The bracket loop, corkscrew loop, and central helix together form a network of hydrophilic and hydrophobic interactions involving both the main chain and amino acid side chains (Fig. 2C). The remaining protomer-protomer contacts occur between a small α helix extending from the bottom of the central helix on one protomer and the central helix of the adjacent protomer (Fig. 2C).
Fig. 2. The trimeric interface of prefusion RABV-G.
(A and B) Book-end models of the interface between protomers of RABV-G (A) and VSV-G (B) (Protein Data Bank ID: 5i2s) with corresponding contacts from adjacent subunits shaded (left and middle). The yellow dashed line indicates the central helix. On the right, the RABV-G and VSV-G central helices and distances between side chains are shown. (C and D) Magnified view of the RABV-G (C) and VSV-G (D) trimeric interface, with the corkscrew loop (yellow), the bracket loop (orange), and the small helix (purple). Dashed black lines denote hydrogen bonds and hydrophobic interactions.
In this structure, the bracket loop and C terminus adopt significantly different conformations compared to a previously reported structure of monomeric RABV-G, in which the corkscrew loop is unresolved (fig. S3) (7). In the monomeric structure, residues 373 to 389 of the bracket loop instead form a helix, which extends into a long, flexible loop that projects sideways and makes crystal contacts with another monomer (fig. S3) (7). This extended conformation disrupts the trimeric interface. The alternative conformations of the bracket and corkscrew loops suggest that the trimeric interface is unstable, which could explain why the trimeric RABV-G structure has been so elusive.
To better understand the trimeric interface, we examined vesicular stomatitis virus (VSV), another member of the family Rhabdoviridae and the most closely related virus to rabies for which a trimeric glycoprotein structure is available (20, 21). The overall organization of RABV-G and VSV-G is similar, but the relatively low sequence identity (~22%) leads to differences in interactions at the trimeric interface. In RABV-G, the central helices are arranged as a cone with a wide top and narrow bottom (Fig. 2A). In contrast, in VSV-G, the central helices form a cone with a narrow top and wide bottom (Fig. 2B), and the glycoprotein assembles primarily through interactions between the central helices, with the bracket loop shifted and forming backbone-backbone hydrogen bonds with the adjacent protomer (Fig. 2D).
Differences in residue composition between RABV-G and VSV-G also influence propensity to trimerize and trimer stability. During the transition to the postfusion conformation, the VSV-G central helices elongate at the top, introducing additional contact points between protomers (21). As a result, VSV-G trimer stability increases with decreasing pH, which favors the transition to the postfusion conformation (21, 22). In RABV-G, where the prefusion central helices are arranged into a cone with a wide top, central helix elongation in the postfusion transition would not be expected to yield strong interprotomer contacts. RABV-G soluble ectodomain trimers destabilize with decreasing pH, as evidenced by a fourfold lower intermonomer binding affinity at pH 5.5 than at 7.4 (fig. S4). Under cryo-EM conditions at pH 5.5, our soluble RABV-G ectodomain has a monomeric and heterogeneous conformation, and an alternate intermediate postfusion conformation RABV-G, unlike postfusion VSV-G, was crystallized as a monomer (7). These findings support our structural evidence that the prefusion trimeric interface of RABV-G is built not primarily by a central core of tightly interacting helices and hydrophobic contacts but instead by lateral interactions between the central helix and adjacent loops.
Neutralizing antibody recognition
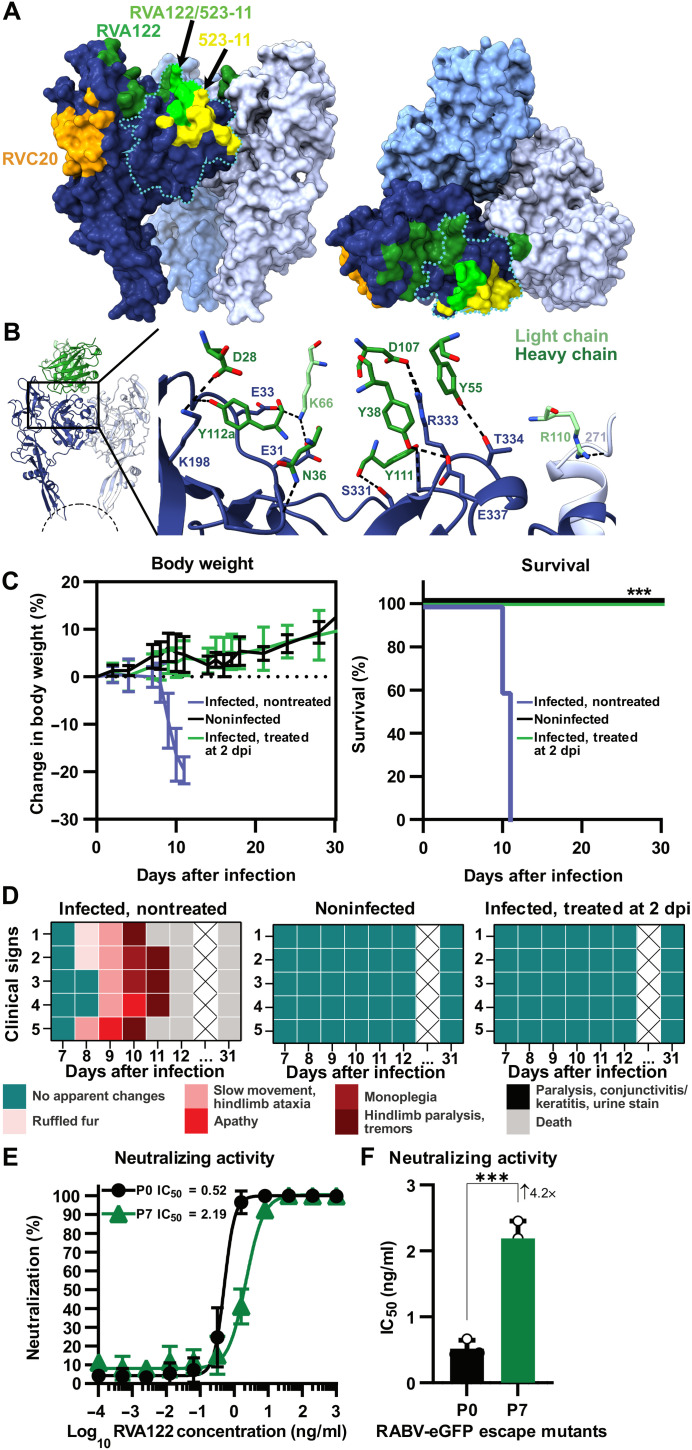
Challenges in developing rabies vaccines that elicit longer lasting immunity include the lack of the quality and durability of the resulting neutralizing antibody response, and the lack of efficacy of most antibodies against other emerging and circulating lyssaviruses (22, 23). Monoclonal antibody RVA122 is the type of antibody desired from rabies immunization. RVA122 potently neutralizes rabies virus with an 90% inhibitory concentration (IC90) of ~0.1 ng/ml and neutralizes multiple other phylogroup I lyssaviruses with similar efficacy (8). Three copies of RVA122 bind to the top of the RABV-G trimer, with each Fab adhering to a quaternary epitope that bridges domains I and III and contacts domain II on an adjacent protomer (Fig. 3). This footprint is unique compared to the two other RABV-G binding antibodies with known structures: RVA20 and 523-11. Both RVA20 and 523-11 neutralize rabies virus, but each binds to a single domain that has the same conformation in pre- and postfusion RABV-G (Fig. 3A) (6, 7).
Fig. 3. RVA122 binds a quaternary epitope.
(A) Binding footprints for antibodies RVA122 (dark green), 523-11 (yellow), and RVA20 (orange), and overlap in footprints between RVA122 and 523-11 (bright green). RABV-G domain I is outlined in a teal dotted line. (B) Interactions between RVA122 (green) and RABV-G (dark and light blue). RVA122 residues are labeled in IMGT (international immunogenetics database) format. (C) Body weight change (left) and survival (right) of mice that were either not infected with RABV, infected with RABV, or infected with RABV and treated with RVA122 at 2 days after infection (n = 5 per group). Body weight data are displayed as means ± SD, and survival is displayed as cumulative Kaplan-Meier survival curves of mice from different experimental groups. A log-rank (Mantel-Cox) test compares treated and nontreated groups (right). (D) Clinical signs of RABV infection in mice that were either not infected with RABV, infected with RABV, or infected with RABV and treated with RVA122 at 2 days after infection. Heatmaps depict a progressive clinical score scale (0: no apparent changes; 1: ruffled fur; 2: slow movement, hindlimb ataxia; 3: apathy; 4: monoplegia; 5: hindlimb paralysis, tremors; 6: paralysis, conjunctivitis/keratitis, urine staining of the haircoat of the perineum; 7: death), where each line represents one animal. (E and F) Neutralization titer of RVA122 on wild-type RABV-Tha–enhanced GFP (eGFP) and escape-mutant RABV-Tha-eGFP (RABV-Tha-eGFP passaged seven times in the presence of RVA122) on BSR cells at 48 hours postinfection. Neutralization curves (E) were generated by fitting data points using a variable slope and a four-parameter regression curve (best-fit method), and median inhibitory concentration (IC50) data (F) were analyzed via the unpaired t test and are displayed as means ± SD. Three independent replicates were performed. ***P < 0.001.
RVA122 binding increases the proportion of RABV-G trimers visible via cryo-EM by over 30-fold, making high-resolution reconstruction possible, and locks RABV-G into the prefusion conformation. RVA122 likely neutralizes rabies virus by inhibiting the transition to the postfusion conformation, as the antibody remains bound below pH 5 after negative staining, and may also block receptor binding, as the RVA122 binding footprint and at least one of the rabies virus receptors overlap (Fig. 3 and fig. S2). Residue contacts between RVA122 and RABV-G include domain I residues S331, R333, T334, and E337 and domain III residues E31, E33, and K198, most of which are highly conserved among phylogroup I lyssaviruses (fig. S7), explaining why RVA122 is broadly neutralizing. RVA122 light-chain residue R110 forms a single contact with domain II residue L271 on the neighboring protomer (Fig. 3B). Mutation of R110 to Ala or Glu did not significantly affect binding affinity; this contact, therefore, does not appear to be critical (fig. S8). Because mutation of RVA122 residue R110 has a negligible effect on antibody binding, the enhanced trimerization of RABV-G in complex with RVA122 likely results from RVA122 stabilizing the prefusion conformation by bridging domains I and III, rather than bridging protomers.
RVA122 is fully protective against rabies virus challenge in mice. Mice treated with RVA122 at a dose of 20 mg/kg at day 2 after infection were completely protected against a challenge with a lethal dose of rabies virus (Fig. 3C) and displayed none of the clinical symptoms associated with infection (Fig. 3D). In contrast, untreated mice all died by day 11 after displaying multiple clinical symptoms associated with rabies infection (Fig. 3D).
When RABV was passaged in vitro in the presence of RVA122, an escape mutant carrying the RABV-G point mutations P137S/R333Q arose after seven passages (fig. S8). The P137S/R333Q escape mutant had a 4.2-fold higher RVA122 median inhibitory concentration (IC50) titer compared to the wild-type virus (Fig. 3, E and F) but was still neutralized by RVA122 at relatively low concentrations of antibody. RABV-G residue P137 is located in domain IV, far from the RVA122 binding site, and it is unclear whether it plays a role in RVA122 binding. Residue R333, however, forms a hydrogen bond with RVA122 heavy-chain residue D107 (Fig. 3B) and has been extensively described for its role as an antigenic site III escape mutation (23, 24). Mutations to R333 (including R333S) have also been reported to attenuate rabies virulence, and recombinant viruses containing R333 mutations have been widely used in the oral vaccination of wildlife (23–25).
Fusion loops stabilize soluble ectodomain trimers
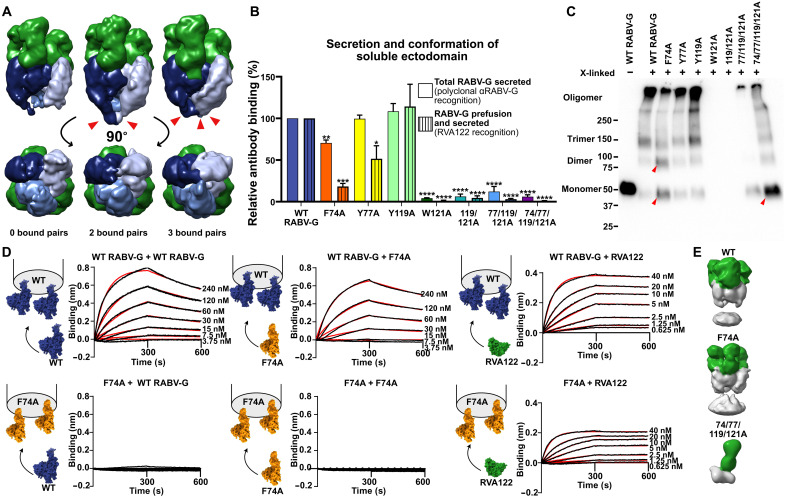
In addition to RVA122 and the trimeric interface, RABV-G fusion loops also appear to play a role in trimerization and to influence protein stability. In a prior study, when RABV-G fusion loops were replaced with Gly-Ser linkers to facilitate protein purification and crystallization, no trimerization was observed at neutral or acidic pH (7). In contrast, we observe that wild-type RABV-G ectodomains containing intact fusion loops associate readily (fig. S4). We also observe that when RABV-G trimers are not anchored to micelles or cellular membranes, the fusion loops are either disordered or interacting with each other (Figs. 1C and 4A). These results suggest that the fusion loops affect glycoprotein ectodomain association and trimerization in addition to driving fusion of the viral and cell membranes after endosomal acidification.
Fig. 4. Fusion loop point mutations affect secretion, conformation, and trimerization of soluble RABV-G ectodomain.
(A) Low-resolution cryo-EM reconstructions of RABV-G/RVA122 complexes with 0, 2, or 3 pairs (red arrowheads) of fusion loops interacting with each other. (B) Enzyme-linked immunosorbent assay (ELISA) quantifying secreted RABV-G and prefusion, secreted RABV-G for wild-type (WT) and mutant soluble ectodomains. Statistical significance of differences between groups was analyzed using analysis of variance (ANOVA). Error bars indicate SEM for three biological replicates. (C) Cross-linking assay showing monomers, dimers, trimers, and higher-order oligomers formed from soluble RABV-G ectodomains. Red arrowheads indicate changes in oligomerization for F74A and 74/77/119/121A mutants compared to the WT. (D) Biolayer interferometry experiments detailing binding interactions between WT RABV-G protomers, F74A RABV-G protomers, and RVA122 Fabs. Curves were fitted (red) using the Octet Data Analysis HT software version 11, with global fitting and a 1:1 fitting model. Experiments were performed in duplicate. (E) Negative stain EM reconstructions of soluble RABV-G (gray) immunoprecipitated with RVA122 Fabs (green). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To evaluate this hypothesis, we made alanine substitutions at aromatic fusion loop residues embedded in membranes and micelles (F74, Y77, Y119, and W121) both individually and in combination and expressed these mutants as soluble ectodomains. We evaluated secretion and conformation of these mutants via enzyme-linked immunosorbent assay (ELISA) and oligomerization via Western blot and negative stain EM. All fusion loop mutations except for Y119A significantly reduce the amount of secreted, prefusion RABV-G ectodomain compared to the wild type (Fig. 4B). F74A results in a 30% reduction in total secreted RABV-G and 80% reduction in prefusion secreted RABV-G (Fig. 4B), whereas Y77A results in a 50% reduction of only secreted prefusion RABV-G. Mutation of W121 to alanine, alone or in combination with any other mutation, severely reduces or eliminates total glycoprotein secretion (Fig. 4B). Expression of W121A-containing constructs could be detected in cell lysates (fig. S5), showing that the decreased secretion is not due to lower protein expression.
In addition to decreasing secretion of the soluble ectodomain, F74A also substantially reduces the amount of trimers, but not dimers, detected via protein cross-linking (Fig. 4C). In biolayer interferometry experiments, F74A mutants bind RVA122 Fabs, indicating that some of the glycoprotein is in the prefusion conformation, but protomers do not stably associate with each other (Fig. 4D). F74A ectodomains immunoprecipitated and stabilized with RVA122, however, do trimerize (Fig. 4E). F74A protomers bind to wild-type RABV-G protomers in biolayer interferometry experiments when wild-type RABV-G is the ligand (attached to the biosensor), but not the analyte (in solution) (Fig. 4D). These results suggest that F74A forms transient dimers in solution that can pair with wild-type protomers to form trimers and that RVA122-mediated stabilization of the prefusion conformation can also overcome the barrier to trimerization.
Soluble ectodomains containing the W121A mutation were poorly secreted, and sufficient amounts of protein could not be produced for biolayer interferometry experiments. W121A, 119/121A, and 77/119/121A were also too poorly secreted to be evaluated in cross-linking experiments. The quadruple mutant 74/77/119/121A forms primarily monomers in cross-linking experiments (Fig. 4C), and 74/77/119/121A soluble ectodomains immunoprecipitated with RVA122 also appear entirely monomeric via negative stain EM (Fig. 4E), indicating that any dimers or trimers that this mutant does form are unstable.
Fusion loops stabilize full-length RABV-G prefusion conformation
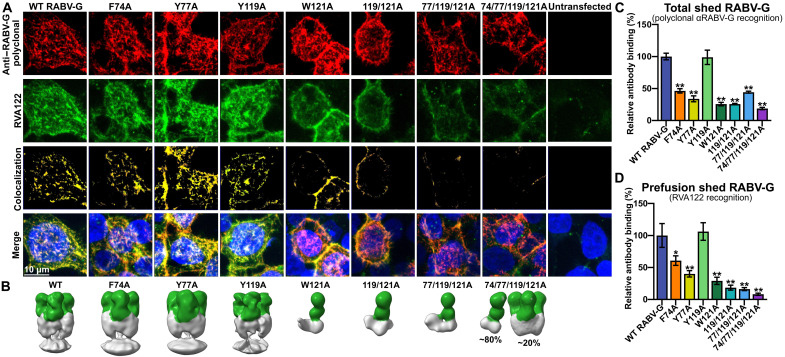
To determine whether the fusion loops also affect the conformation and stability of full-length RABV-G, we expressed the fusion loop point mutants as full-length glycoproteins, which include a transmembrane domain and cytoplasmic tail, and analyzed them with flow cytometry and immunofluorescence staining. All full-length versions of the RABV-G mutants express well and readily reach the cell surface (Fig. 5A). However, for mutants containing W121A, significantly less of the full-length protein is in the prefusion conformation (fig. S6), and the prefusion RABV-G localizes to different areas of the cell compared to the wild type (Fig. 5A).
Fig. 5. Fusion loop mutations affect full-length RABV-G conformation and trimerization.
(A) Confocal immunofluorescence of cell surface-displayed, full-length RABV-G with fusion loop point mutations, stained for total RABV-G expression (polyclonal) and prefusion RABV-G (RVA122). Images are displayed as maximum intensity projections. (B) Negative stain EM reconstructions of shed RABV-G (gray) purified via immunoprecipitation with RVA122 (green). (C and D) ELISA quantification of the amount of total shed RABV-G (C) or prefusion shed RABV-G (D) carrying fusion loop mutations relative to the WT. Statistical significance of differences between groups was analyzed using ANOVA. Error bars indicate SEM for three biological replicates. *P < 0.05; **P < 0.01.
In rhabdoviruses, the full-length glycoprotein can be either membrane-anchored or proteolytically cleaved near the transmembrane domain, releasing a shed ectodomain (26–28). Shed RABV-G is slightly longer than the cloned, soluble ectodomain (447 amino acids instead of 420) and contains no purification tags (28). The role of shed RABV-G during infection is unknown, but it may serve as a decoy for the immune system, as has been observed for other viruses (29–31). Furthermore, shed VSV-G has been reported to bind to full-length glycoprotein for uptake into cells (26) and injection of soluble RABV-G ectodomains into the brains of mice increases locomotion (32), a rabies symptom believed to facilitate transmission to new hosts.
We observe that fusion loop mutations affect the release of shed glycoprotein into tissue culture supernatant. All point mutations except Y119A decrease the total amount of shed RABV-G relative to the wild type (Fig. 5C). Furthermore, there is also a significant decrease in the ratio of prefusion shed RABV-G to total shed RABV-G for the 77/119/121A and 74/77/119/121A mutants (fig. S6). To visualize the structure of shed RABV-G, we immunoprecipitated mutants with RVA122 and examined them with negative stain EM. Shed wild-type, F74A, Y77A, and Y119A RABV-G each form trimers when in complex with RVA122, with the fusion loops embedded in cellular membranes that coprecipitate with the glycoprotein (Fig. 5B). In contrast, none of the mutants that contain W121A copurify with cellular membranes. They are also mostly or entirely monomeric, with ~20% of the RVA122-captured 74/77/119/121A RABV-G–forming trimers and the other mutants containing W121A-forming monomers (Fig. 5B).
Together, these results suggest that for the full-length glycoprotein, W121 stabilizes the prefusion conformation by interacting with membranes, preventing the glycoprotein from shifting into alternate conformations. Interactions between W121 and the viral membrane also likely stabilize RABV-G on the surface of virions, as has been observed for Mokola virus glycoprotein (33). The ability of shed 74/77/119/121A to trimerize in complex with RVA122 but not the other W121A mutants or 74/77/119/121A soluble ectodomain (Fig. 4E) is puzzling but might be explained by relative protein expression levels or interactions among the fusion loops.
DISCUSSION
Rabies virus infection is more than 99% lethal, still kills more than 50,000 people annually, and is endemic in all populated continents. RABV-G is a critical component of vaccines and target for antibody therapeutics and potential antiviral drugs. Despite considerable advances in the fields of immunology and vaccinology, rabies vaccines do not yet elicit lifelong immunity, leaving many individuals with no detectable rabies antibodies within 1 year of vaccination. Structural heterogeneity of the RABV-G likely contributes to the suboptimal antibody response. Prefusion, trimeric RABV-G would be an ideal immunogen for vaccination, but presentation of this form has been challenging due to the inherent instability of the molecule. Lack of information on what the trimeric structure is, how it is organized, and how it can be stabilized has hampered these efforts.
Here, we report the structure of trimeric, prefusion RABV-G bound to a potent and broadly neutralizing antibody. In this structure, the fusion loops are anchored into cellular membrane/micelle density, a feature not appreciated in a previously published structure of the alternate prefusion intermediate conformation RABV-G monomer, in which the fusion loops were replaced by linkers. We show that the fusion loops play an integral role in stabilizing the prefusion conformation, regardless of whether RABV-G is full-length, shed, or expressed as a soluble ectodomain. Hence, inclusion of these elements in vaccine designs will be critical for the faithful display of the prefusion RABV-G trimer. A nucleic acid–based vaccine may bypass issues associated with purification but must still direct expression of glycoproteins with the correct conformation and quaternary assembly.
While this manuscript was under revision, we became aware of a bioRxiv preprint describing a similar structure of trimeric RABV-G in complex with a pair of neutralizing antibodies targeting different antigenic sites (34). This work provides additional information on RABV-G/antibody interactions that should prove useful in the development of rabies antibody therapeutics and corroborates our model of the RABV-G trimeric interface.
The prefusion, trimeric RABV-G structure illustrated here, the demonstration of the importance of fusion loops for successful trimerization and conformational stabilization, and the visualization of the potent and broadly neutralizing antibody epitope together provide a key road map to guide the development of improved rabies vaccines and postexposure therapeutics.
MATERIALS AND METHODS
Experimental design
This study was designed to determine the structure of prefusion RABV-G trimer, understand how the RABV-G fusion loops affect protein conformation and oligomerization, and examine the interactions of RABV and the RABV-G trimer with a potently neutralizing antibody. Cryo-EM was used to determine the structure of the RABV-G prefusion trimer in complex with the neutralizing antibody. In optimizing RABV-G constructs and freezing conditions for cryo-EM, we identified the RABV-G fusion loops as a possible source of stability for the prefusion trimer. To test this hypothesis, we subsequently performed a range of biochemical experiments including protein cross-linking, biolayer interferometry, negative stain EM, immunofluorescence assays, and ELISAs. To examine the interaction between RABV-G and the potently neutralizing antibody RVA122, we also performed in vivo neutralization assays, in vitro neutralization assays, and biolayer interferometry.
Cells
Fabs were prepared in S2 insect cells (Drosophila melanogaster; Invitrogen, no. R69007) grown in Insect-XPRESS medium (Lonza) with 1% penicillin-streptomycin (Thermo Fisher Scientific) at 27°C in a rotary shaker. RABV-G ectodomain and full-length protein were produced in human embryonic kidney (HEK) 293T cells (Homo sapiens; American Type Culture Collection, no. CRL-3216, RRID:CVCL_0063) grown in T75 flasks (Corning) in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Thermo Fisher Scientific) at 37°C with 5% CO2. Viral titration and neutralization assays were performed using BSR cells (a BHK-21 clone, provided by M. Lafon, Institut Pasteur, Paris) grown in DMEM with 10% FBS.
RABV strains and titration
Tha–green fluorescent protein (GFP) recombinant virus (EVAg collection Ref-SKU: 014 V-03195) is based on the wild isolate 8743THA (EVAg collection Ref-SKU: 014 V-02106), isolated from a human bitten by a dog in 1983 in Thailand. Titrations were performed on BSR cells using the fluorescent focus method (35). A total of 20 μl of serial dilutions (1 to 5) of virus were inoculated in duplicates on 5 × 104 BSR cells and incubated at 37°C. At 40 hours after infection, the number of GFP foci was determined under a fluorescent microscope, and the titer was calculated in fluorescent focus units per milliliter (FFU/ml).
Plasmids and cloning
Codon-optimized sequences for the heavy and light variable domains of the neutralizing antibody RVA122 were synthesized and cloned into pMT puro and pMT plasmids, respectively, containing the corresponding regions for Fab immunoglobulin G1 heavy and immunoglobulin K light chains. The heavy-chain sequence also contained a C-terminal enterokinase cleavage site, followed by a double-strep tag.
The codon-optimized sequence for full-length, PV strain RABV-G (National Center for Biotechnology Information reference sequence: NC_001542.1) was synthesized and cloned into the pcDNA3.1(−) vector. The soluble ectodomain was cloned by truncating the full-length glycoprotein to residues 1 to 439 (numbering includes signal peptide) and adding either no tag, a V5/6x-His tag, or a double strep-tag and an avi-tag to the C terminus. Point mutations to full-length and soluble ectodomain plasmids were made via site-directed mutagenesis using Q5 DNA polymerase [New England Biolabs (NEB)], T4 polynucleotide kinase (NEB), and T4 DNA ligase (NEB).
Protein expression and purification
For RVA122 expression, heavy-chain and light-chain plasmids were cotransfected into S2 cells using Effectene (QIAGEN) according to the manufacturer’s protocol and selected using puromycin (Invivogen). Cells were expanded after selection, transferred to shaker flasks (TriForest Enterprises), and induced with CuSO4 (500 μM) and sodium butyrate (5 mM) once cells had reached a density of ~1 × 107 cells/cm2. Cells were harvested 4 days after induction and pelleted at 4000g. Supernatant was saved and adjusted to pH 8.0 with NaOH and then freeze-thawed at −20°C/25°C to precipitate salts from the supernatant before purification. Supernatant was filtered, and Fabs were purified on a StrepTrap HP column (GE Healthcare). Eluted fractions were pooled and concentrated using a 10-kDa molecular weight cutoff (MWCO) centrifugal filter (Millipore), followed by further fractionation over an S200i column (GE Healthcare) in phosphate-buffered saline (PBS) at 0.5 ml/min to remove aggregates.
For RABV-G ectodomain expression, HEK 293T cells seeded in T75 flasks (Corning) at a density of 4 × 104 cells/cm2 were transiently transfected with 9.8 μg of the recombinant plasmid using polyethyleneimine (PEI) at a 3:1 PEI:DNA ratio. At 2 and 4 days after transfection, supernatant was collected from flasks and centrifuged at 4000g to remove cell debris before purification. Tagged RABV-G ectodomain was purified from supernatant with Ni–nitrilotriacetic acid (NTA) agarose beads (QIAGEN) for His-tagged protein or Strep-Tactin Superflow plus beads (QIAGEN) for strep-tagged protein. Untagged RABV-G ectodomain was purified from supernatant via immunoprecipitation with the strep-tagged RVA122 Fab on Strep-Tactin Superflow plus beads (QIAGEN).
Briefly, the supernatant was adjusted to pH 8.0 with NaOH and then incubated overnight at 4°C with either beads for tagged RABV-G ectodomain or with RVA122 Fab-captured beads. Beads were pelleted from supernatant via centrifugation, washed three times with PBS, and then eluted multiple times with either 250 mM imidazole/tris-buffered saline for Ni-NTA beads or 10 mM d-desthiobiotin/PBS for Strep-Tactin beads. Protein for cryo-EM was concentrated and buffer-exchanged into PBS in a 100-kDa MWCO centrifugal filter (Millipore), which removed unbound RVA122 Fabs.
Shed RABV-G was produced by expression of full-length RABV-G in 293T cells and collection of supernatant 4 days after transfection. Purified, shed RABV-G was produced via immunoprecipitation with RVA122 Fabs, as described above.
Preparation of cryo-EM grids and data collection
Purified RABV-G/RVA122 complexes at a concentration of ~200 μg/ml were mixed 3:1 with 0.12 mM (0.03 mM final concentration) lauryl maltose neopentyl glycol (LMNG; Anatrace) and immediately frozen on 1.2-/1.3-μm C-flat grids [Electron Microscopy Sciences (EMS)] using an FEI vitrobot Mark IV (Thermo Fisher Scientific) in 85% humidity and 4°C with a 10-s blot time. A total of 1969 micrographs were collected on a 200-keV Talos Arctica with a K2 direct electron detector (Gatan) at the Pacific Northwest Cryo-EM Center (PNCC) with pixel size of 1.15 Å/pixel and a total dose of 26.7 e−/Å2.
A second cryo-EM dataset of 6305 micrographs was collected with purified RABV-G/RVA122 complexes (~200 μg/ml) mixed 3:1 with 0.36 mM (0.09 mM final concentration) LMNG and frozen on 2-/1-μm C-flat grids in 85% humidity and 4°C with a 10-s blot time. Images were collected on a 300-keV Titan Krios electron microscope with a K3 direct electron detector at 1.1 Å/pixel and a total dose of 50.0 e−/Å2.
Cryo-EM data processing and model building
The dataset containing RVA122/RABV-G complexes with 0.09 mM LMNG (table S1) was used for high-resolution reconstruction and model building. Recorded movies were motion-corrected and dose-weighed using either cryoSPARC’s own algorithm or RELION’s MotionCorr2 implementation. Contrast transfer function (CTF) parameters were determined either with cryoSPARC’s Patch CTF program or in RELION using CTFFIND4 (36).
Particles were picked using the TOPAZ neural network picker (37) and processed in cryoSPARC (38). Two-dimensional (2D) classification and 3D hetero-refinement were used to screen for particles bound to cellular membrane or micelle densities. Symmetry-expanded particles were used in a local refinement job to produce the first high-resolution map. To further improve the map quality around the fusion loops, particle coordinates were transferred to RELION 3.1 (39). In RELION, particles were sorted via 3D classification without alignment to isolate complexes with better resolved fusion loops. Per-particle CTF parameters were estimated, and particles were Bayesian-polished to yield the final map. Final half-maps were transferred to cryoSPARC to estimate the local resolution. A detailed flowchart containing data processing steps is presented in fig. S9.
Model building was performed in Coot 0.9.2 (40) and ISOLDE (41). Model refinement and validation was performed in Phenix 1.19 (42). Model building was aided by a protein model generated from AlphaFold2 (43, 44). ChimeraX 1.2.5 (45) was used to prepare figures of the structure. Fourier shell correlation curves were generated using RELION, the local resolution histogram using cryoSPARC, and correlation coefficient graphs using Phenix.
Lower-resolution cryo-EM maps of RABV-G/RVA122 complexes not bound to micelles or cellular membranes were prepared from the dataset containing complexes with 0.03 mM LMNG. Particles were picked with TOPAZ, as above, reconstructed into an initial model in cryoSPARC with C3 symmetry, and then sorted via 3D variability analysis focusing on the fusion loops to identify particles with fusion loops in different conformations. Groups of similar particles were reconstructed with a homogeneous refinement job and C1 symmetry.
Negative stain EM and reconstruction
Shed RABV-G and soluble RABV-G were purified via immunoprecipitation with RVA122 Fabs, as described above, then added to C-flat CF400Cu grids (EMS) at a concentration of approximately 10 μg/ml, and stained with 0.5% uranyl acetate. Grids were imaged on a Titan Halo electron microscope operated at 300 keV with a Falcon 3 direct electron detector at 1.87 pixels/Å and 50 e−/Å2. Particles were picked with TOPAZ and reconstructed in Cryosparc using homogeneous refinement and C3 symmetry.
Enzyme-linked immunosorbent assays
HEK293T cells were seeded in six-well plates (Corning) at a density of 4 × 104 cells/cm2 and grown overnight. Cells were then transfected with plasmids encoding full-length or soluble rabies glycoprotein using PEI at a ratio of 1:3 DNA to PEI (1.26 μg of DNA and 3.8 μg of PEI per well). Four days after transfection, the supernatant was removed from cells, diluted 1:1 in PBS, then transferred to half-well ELISA plates (Corning), and incubated for 1 hour at room temperature. The supernatant was removed and wells were blocked with 3% bovine serum albumin (BSA; Sigma-Aldrich) in PBS for an additional hour at room temperature. Wells were washed once with PBS with 0.05% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) (BioVision) and then incubated with either 1:2000 rabbit anti–RABV-G polyclonal antibody (a gift of M. Schnell, Thomas Jefferson University) or RVA122 monoclonal antibody for 1 hour at room temperature. Plates were washed three times with PBS/0.05% CHAPS and then incubated with either 1:2000 goat anti-human Fab horseradish peroxidase (HRP; Jackson ImmunoResearch Laboratories, catalog no. 109-036-006, RRID:AB_2337590) or 1:2000 goat anti-rabbit HRP (SouthernBiotech, catalog no. 4050-05, RRID:AB_2795955) for 1 hour at room temperature. Plates were washed three times with PBS/0.05% CHAPS, then developed with 1-step Ultra TMB-ELISA substrate solution (Thermo Fisher Scientific), and quenched with 1 M sulfuric acid (Thermo Fisher Scientific). Plates were read at 450 nm on a Tecan Spark 10M plate reader. Three biological replicates were performed for each experiment, and results were analyzed via analysis of variance (ANOVA) in GraphPad Prism 9.
Protein cross-linking assays
Purified His-tagged RABV-G ectodomains were buffer-exchanged into PBS, diluted to a concentration of 56 μg/ml, and incubated in 0.01% glutaraldehyde/PBS for 1 hour at room temperature. Cross-linking was quenched via addition of 1 M tris (pH 8.0) to a final concentration of 50 mM. Proteins were separated via SDS–polyacrylamide gel electrophoresis (PAGE) gel under reducing conditions, transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore), and stained with 1:2000 rabbit polyclonal anti–RABV-G antibody and 1:2000 goat anti-rabbit HRP antibody (SouthernBiotech, catalog no. 4050-05, RRID:AB_2795955) for visualization.
To determine whether the glutaraldehyde cross-linker resulted in protein aggregation, disuccinimidyl suberate (DSS; Thermo Fisher Scientific) cross-linking was also tested. Purified, wild-type RABV-G ectodomains were incubated with increasing concentrations of DSS according to the manufacturer’s protocol, covering the entire range of recommended concentrations, from 0.25 to 5 mM. Proteins were incubated with DSS for 30 min at room temperature, followed by addition of 1 M tris (pH 8.0) to a final concentration of 50 mM to quench the reaction, and incubation for an additional 15 min at room temperature. Proteins were visualized via Western blotting, as described above (fig. S5).
Measurement of mutant RABV-G ectodomain expression levels
293T cells seeded in six-well plates at a density of 4 × 104 cells/cm2 were transfected with mutant RABV-G ectodomains as described above. Four days after transfection, cells were washed with PBS; lysed with 1% Triton X-100 in PBS; separated via SDS-PAGE gel under reducing conditions; transferred to a PVDF membrane; and stained with 1:2000 rabbit polyclonal anti–RABV-G antibody and 1:2000 mouse anti–β-actin antibody (Santa Cruz Biotechnology, no. sc-69879, RRID:AB_1119529), then 1:2000 goat anti-mouse HRP antibody (Thermo Fisher Scientific, catalog no. 31437, RRID:AB_228295), and 1:2000 goat anti-rabbit HRP antibody (SouthernBiotech, catalog no. 4050-05, RRID:AB_2795955) for visualization.
Immunofluorescence assays and flow cytometry
HEK293T cells were seeded on 12-mm circular glass coverslips (Fisher) in a 24-well plate (Corning) at a density of 4 × 104 cells/cm2 and grown overnight. Cells were then transfected with plasmids encoding full-length RABV-G using PEI at a ratio of 1:3 DNA to PEI (0.25 μg of DNA and 0.75 μg of PEI per well). Two days after transfection, cells were fixed in 4% paraformaldehyde (PFA)/PBS for 10 min at room temperature and then stained with 1:2000 rabbit polyclonal anti–RABV-G and 1:2000 RVA122 monoclonal antibody in PBS with 0.1% BSA for 1 hour at room temperature. Cells were washed twice with PBS and then stained with 1:2000 anti-human fluorescein isothiocyanate (FITC; SouthernBiotech, catalog no 2040-02, RRID:AB_2795641) and 1:2000 anti-rabbit Alexa Fluor 568 (Thermo Fisher Scientific, catalog no. A-11011, RRID:AB_143157) for 1 hour at room temperature. Cells were washed twice with PBS and then stained with 1:10,000 Hoechst (10 mg/ml) for 15 min and washed with PBS. Coverslips were mounted on slides with ProLong Gold Antifade Mountant (Thermo Fisher Scientific) and imaged on a Zeiss 880 confocal microscope at ×40 magnification.
For flow cytometry experiments, HEK293T cells were similarly seeded in six-well plates and transfected with plasmids encoding full-length RABV-G. Two days after transfection, cells were fixed with 4% PFA, washed with PBS, incubated in Human BD Fc Block (BD Biosciences), and stained with 1:1000 anti–RABV-G polyclonal antibody and 1:1000 RVA122 monoclonal antibody for 1 hour. Cells were washed twice and then stained with 1:1000 goat anti-rabbit Alexa Fluor 647 (Thermo Fisher Scientific, catalog no. A27040, RRID:AB_2536101) and 1:1000 goat anti-human FITC (SouthernBiotech, catalog no. 2040-02, RRID:AB_2795641) for 1 hour. Cells were washed twice and then quantified using a LSR II flow cytometer (BD Biosciences). Experiments were performed in triplicate (biological replicates), with untransfected cells serving as a negative control. RVA122 mean fluorescence intensity was analyzed via ANOVA in GraphPad Prism 9.
Biolayer interferometry
RABV-G ectodomains used to quantify binding kinetics contained a C-terminal double strep tag and avi-tag. RABV-G used as a ligand was biotinylated at the avi-tag using BirA ligase according to the manufacturer’s protocol (Avidity) and then buffer-exchanged into PBS to remove unlinked biotin. Binding kinetics were measured using an Octet Red 384 (ForteBio). Streptavidin biosensors (ForteBio) were hydrated in kinetics buffer [PBS (pH 7.4) with 0.1% BSA and 0.02% CHAPS] for 10 min at room temperature. Baseline readings were collected in kinetics buffer for 30 s at 30°C, followed by loading of biotinylated RABV-G onto biosensors at a concentration of ~3.5 μg/ml for 300 s and a 60-s wash in kinetics buffer. For measurements of Fab binding affinity, RVA122 wild-type and mutant Fabs at concentrations ranging from 1.25 to 80 nM were incubated with biosensors for 300 s to measure association, followed by a 300-s incubation in kinetics buffer to measure disassociation. For measurements of RABV-G binding affinity, biosensors were incubated with unbiotinylated RABV-G ectodomain at concentrations ranging from 3.75 to 960 nM, followed by a 300-s incubation in kinetics buffer. For measurements at pH 5.5, all steps were performed in 0.1 M citric acid/trisodium citrate buffer (pH 5.5) with 0.1% BSA and 0.02% CHAPS. Experiments were performed in duplicate (technical replicates), and a sensor run without analyte was used as a reference. Affinity rate constants were calculated by subtracting the reference and then applying global fitting using the Octet Data Analysis HT software version 11 (ForteBio) and a 1:1 fitting model. In one case (fig. S4B), the 1:1 binding model yielded a poor fit of the experimental data. For this dataset, we also applied a 2:1 fitting model and showed both 1:1 and 2:1 models.
RVA122 protection of RABV-infected mice
The murine rabies model was based on (46), with a few modifications. Briefly, 8-week-old female specific pathogen–free (SPF) Balb/cJRj mice were purchased from Janvier Laboratories and handled under SPF conditions, according to the institutional guidelines of the Central Animal Facility at Institut Pasteur, with ad libitum access to water and food. Before any manipulation, animals underwent an acclimation period of 1 week. Animals were infected with 4000 FFU of the pathogenic Tha-RABV strain (isolate 8743THA, EVA Global collection, Ref-SKU: 014 V-02106) in a total volume of 100 μl, injected into the gastrocnemius muscle of both hindlimbs (two injections of 25 μl in each limb). At 2 days after infection, RABV-infected animals were treated with the monoclonal antibody RVA122 (20 mg/kg) in a final volume of 200 μl per mouse, administered at the site of the infection (two injections of 50 μl in each limb). A total of 15 mice were used and divided into three experimental groups: (i) noninfected and nontreated (n = 5), (ii) infected and nontreated (n = 5), and (iii) infected and treated at 2 days postinfection (dpi) with RVA122 (n = 5). The animals were monitored on a daily basis up to 31 dpi, with body weight and clinical signs recorded. The clinical signs were classified in a progressive 0 to 7 scale (0: no apparent changes; 1: ruffled fur; 2: slow movement, hindlimb ataxia; 3: apathy; 4: monoplegia; 5: hindlimb paralysis, tremors; 6: paralysis, conjunctivitis/keratitis, urine staining of the haircoat of the perineum; 7: death).
Selection of escape mutants resistant to mAb RVA122
To select escape mutants resistant to mAb RVA122, sequential passages of Tha-GFP virus were performed in presence of serial dilutions of RVA122. Eight fivefold serial dilutions of RVA122 were prepared in 12-well plates and incubated for 1 hour at 37°C with 60,000 FFU of Tha-GFP virus [multiplicity of infection (MOI) of 0.1)]. A positive control of infection (virus without RVA122) and a cellular control (without virus) were performed in parallel. After neutralization, 600,000 BSR cells were added and incubated for 4 days at 37°C. Then, viral infection was monitored by microscopy to evaluate the expression of the GFP protein. The virus produced with the higher RVA122 concentration giving a GFP signal comparable to the positive control was used to perform the next passage: Virus was 1:10 diluted and neutralized with the eight fivefold serial dilution of RVA122 as described above. Seven sequential passages were performed. At each passage, supernatants were harvested and stored at −80°C to be used for titration and/or next-generation sequencing.
Virus neutralization test
Virus neutralization tests were performed according to Hellert et al. (6) with slight modifications. A total of 500 FFU of RABV (Tha-GFP or selected RVA122-escape mutants) were incubated with different concentrations of RVA122 in DMEM with 10% FBS in 384-well plates (Greiner Bio-One, no. 781091) for 1 hour at 37°C in a humid atmosphere under 5% CO2. A total of 2500 BSR cells were then added to each well, and the plates were incubated at 37°C (final MOI of 0.2). After 48 hours of incubation at 37°C in a humid atmosphere under 5% CO2, the cells were fixed with 4% PFA and washed in PBS, and the nuclei were counterstained with 20 μM Hoechst 33342. Image acquisitions of four fields per well (6.7 mm2 per well) were performed on the Opera Phenix High Content Screening System (PerkinElmer) using the 10× objective. The percentage of GFP-positive cells was determined using the Harmony High-Content Imaging and Analysis Software (PerkinElmer). IC50 values were determined by nonlinear regression analysis (GraphPad Prism v.9.3.1) from three independent experiments.
Next-generation sequencing
Viral RNAs were extracted from 200 μl of cell culture supernatants recovered at the different passages with TRIzol extraction and Rneasy mini kit (QIAGEN) elution, purified using Agencourt RNAClean XP beads (Beckman Coulter) at a ratio of 1.8, as recommended by the manufacturer, and eluted in 10 μl of nuclease-free water. Eight microliters of the purified RNA were then reverse-transcribed in cDNA using random hexamers (Invitrogen, Illkirch, France) and Superscript III reverse transcriptase (Invitrogen), according to the manufacturer’s instructions. Afterward, double-stranded DNA (dsDNA) was synthesized at 16°C for 2 hours in 80 μl of final reaction mixture, including 20 μl of cDNA, 8 μl of 10× Second Strand Reaction Buffer (NEB, Evry, France), 3 μl of deoxynucleotide triphosphate mix (10 mM; Invitrogen), 1 μl (10 U) of Escherichia coli DNA ligase (NEB), 4 μl (40 U) of E. coli DNA polymerase I (NEB), 1 μl (5 U) of E. coli ribonuclease H (NEB), and 43 μl of nuclease-free water. Last, dsDNA was purified using AMPure XP (Beckman Coulter) at a ratio of 1.8, as recommended by the manufacturer, and eluted in 20 μl of nuclease-free water.
The dsDNA libraries were constructed using the TruSeq DNA PCR-Free libraries prep kit (Illumina, San Diego, USA) and sequenced on Illumina NextSeq 2000 platform. The data obtained were analyzed using the Galaxy@Pasteur software (47): The sequences are first assembled de novo, without any reference sequence, to obtain “contigs,” and then they are aligned on a reference sequence, making it possible to extract the nucleotide differences present in at least 2% of the cases (fig. S8).
Statistical analyses
All statistical analyses were performed using GraphPad Prism version 9.0.0. In vivo neutralization was graphed as a cumulative Kaplan-Meier survival curve, and comparisons between groups were performed using a log-rank (Mantel-Cox) test. In vitro neutralization curves were generated by fitting data points using a variable slope and a four-parameter regression curve (best-fit method), and IC50 values for P0 and P7 virus were compared via the unpaired t test. ELISA and flow cytometry data were analyzed using an ordinary, one-way ANOVA. Numbers of replicates per experiment are indicated in the figure legends. Experiments were represented as means ± SEM or SD, as indicated in the figure legends. P < 0.05 was considered to be statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Acknowledgments
We thank S. Schendel for manuscript editing and L. Kergoat (Lyssavirus Epidemiology and Neuropathology Unit, Institut Pasteur, Paris, France) for technical assistance with virus and mouse experiments. Part of this work was performed at the UtechS Photonic BioImaging (PBI) platform supported by Institut Pasteur and by Région Ile-de-France (program DIM1Health). We also thank G. M. Haustant and L. Lemée (Biomics Platform, C2RT, Institut Pasteur, Paris, France), supported by France Génomique (ANR-10-INBS-09), and IBISA for Illumina sequencing.
Funding: This work was supported by National Institutes of Health (NIH) grants 5T32AI07244-36 (H.M.C.) and 5F32AI147531-03 (H.M.C.) and by Swiss National Science Foundation Early Postdoc Mobility Fellowship P2EZP3_195680 (D.Z.). A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. Confocal microscopy on the Zeiss LSM 880 was supported by equipment grant NIH S10OD021831.
Author contributions: Conceptualization: H.M.C. and E.O.S. Formal analysis: H.M.C., D.Z., F.L., and G.D.d.M. Funding acquisition: E.O.S., H.M.C., D.Z., and H.B. Investigation: H.M.C., R.D.A., A.A., F.L., G.D.d.M., and H.B. Methodology: H.M.C., D.Z., F.L., and G.D.d.M. Resources: D.C. and H.B. Validation: H.M.C., K.M.H., D.Z., F.L., G.D.d.M., and H.B. Visualization: H.M.C., F.L., and G.D.d.M. Writing—original draft: H.M.C. Writing—review and editing: H.M.C., K.M.H., H.B., and E.O.S.
Competing interests: D.C., G.D.d.M., and H.B. hold patents on the use of monoclonal antibodies against rabies virus: PCT/EP2019/078439 (antibodies and methods for the treatment of lyssavirus infection; D.C., G.D.d.M., and H.B.) and PCT/EP2014/003076 (antibodies that potentially neutralize rabies virus and other lyssaviruses and uses thereof; D.C. and H.B.). D.C. is an employee of Vir Biotechnology and may hold shares in Vir Biotechnology. All other authors declare that they have no competing interests.
Data and materials availability: The cryo-EM density maps were deposited in the Electron Microscopy Data Bank under the EMDB accession codes EMD-26397, EMD-26398, EMD-26399, and EMD-26400, and the atomic model was deposited in the Protein Data Bank under the PDB accession code 7U9G. All other material needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Table S1
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.Hampson K., Coudeville L., Lembo T., Sambo M., Kieffer A., Attlan M., Barrat J., Blanton J. D., Briggs D. J., Cleaveland S., Costa P., Freuling C. M., Hiby E., Knopf L., Leanes F., Meslin F.-X., Metlin A., Miranda M. E., Müller T., Nel L. H., Recuenco S., Rupprecht C. E., Schumacher C., Taylor L., Vigilato M. A. N., Zinsstag J., Dushoff J., Global Alliance for Rabies Control Partners for Rabies Prevention, Estimating the global burden of endemic canine rabies. PLoS Negl. Trop. Dis. 9, e0003709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X., Zhu Z., Wang C., Persistence of rabies antibody 5 years after postexposure prophylaxis with vero cell antirabies vaccine and antibody response to a single booster dose. Clin. Vaccine Immunol. 18, 1477–1479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parize P., Sommé J., Schaeffer L., Ribadeau-Dumas F., Benabdelkader S., Durand A., Tarantola A., Cailhol J., Goesch J., Kergoat L., Le Guern A.-S., Mousel M.-L., Dacheux L., Consigny P.-H., Fontanet A., Francuz B., Bourhy H., Systematic booster after rabies pre-exposure prophylaxis to alleviate rabies antibody monitoring in individuals at risk of occupational exposure. Vaccine 9, 309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization , Rabies vaccines: WHO position paper, April 2018–Recommendations. Vaccine 36, 5500–5503 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Fooks A. R., Shipley R., Markotter W., Tordo N., Freuling C. M., Müller T., McElhinney L. M., Banyard A. C., Rupprecht C. E., Renewed public health threat from emerging lyssaviruses. Viruses 13, 1769 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hellert J., Buchrieser J., Larrous F., Minola A., de Melo G. D., Soriaga L., England P., Haouz A., Telenti A., Schwartz O., Corti D., Bourhy H., Rey F. A., Structure of the prefusion-locking broadly neutralizing antibody RVC20 bound to the rabies virus glycoprotein. Nat. Commun. 11, 596 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang F., Lin S., Ye F., Yang J., Qi J., Chen Z., Lin X., Wang J., Yue D., Cheng Y., Chen Z., Chen H., You Y., Zhang Z., Yang Y., Yang M., Sun H., Li Y., Cao Y., Yang S., Wei Y., Gao G. F., Lu G., Structural analysis of rabies virus glycoprotein reveals pH-dependent conformational changes and interactions with a neutralizing antibody. Cell Host Microbe 27, 441–453.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 8.De Benedictis P., Minola A., Rota Nodari E., Aiello R., Zecchin B., Salomoni A., Foglierini M., Agatic G., Vanzetta F., Lavenir R., Lepelletier A., Bentley E., Weiss R., Cattoli G., Capua I., Sallusto F., Wright E., Lanzavecchia A., Bourhy H., Corti D., Development of broad-spectrum human monoclonal antibodies for rabies post-exposure prophylaxis. EMBO Mol. Med. 8, 407–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller T., Dietzschold B., Ertl H., Fooks A. R., Freuling C., Fehlner-Gardiner C., Kliemt J., Meslin F. X., Franka R., Rupprecht C. E., Tordo N., Wanderler A. I., Kieny M. P., Development of a mouse monoclonal antibody cocktail for post-exposure rabies prophylaxis in humans. PLoS Negl. Trop. Dis. 3, e542 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaudin Y., Ruigrok R. W., Tuffereau C., Knossow M., Flamand A., Rabies virus glycoprotein is a trimer. Virology 187, 627–632 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaudin Y., Tuffereau C., Durrer P., Flamand A., Ruigrok R. W., Biological function of the low-pH, fusion-inactive conformation of rabies virus glycoprotein (G): G is transported in a fusion-inactive state-like conformation. J. Virol. 69, 5528–5534 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudin Y., Tuffereau C., Durrer P., Brunner J., Flamand A., Ruigrok R., Rabies virus-induced membrane fusion. Mol. Membr. Biol. 16, 21–31 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Gaudin Y., Raux H., Flamand A., Ruigrok R. W., Identification of amino acids controlling the low-pH-induced conformational change of rabies virus glycoprotein. J. Virol. 70, 7371–7378 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudin Y., Tuffereau C., Segretain D., Knossow M., Flamand A., Reversible conformational changes and fusion activity of rabies virus glycoprotein. J. Virol. 65, 4853–4859 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche S., Gaudin Y., Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology 297, 128–135 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Gaudin Y., Ruigrok R. W., Knossow M., Flamand A., Low-pH conformational changes of rabies virus glycoprotein and their role in membrane fusion. J. Virol. 67, 1365–1372 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rustici M., Santucci A., Lozzi L., Petreni S., Spreafico A., Neri P., Bracci L., Soldani P., A monoclonal antibody to a synthetic fragment of rabies virus glycoprotein binds ligands of the nicotinic cholinergic receptor. J. Mol. Recognit. 2, 51–55 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Tuffereau C., Bénéjean J., Blondel D., Kieffer B., Flamand A., Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 17, 7250–7259 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans J. S., Horton D. L., Easton A. J., Fooks A. R., Banyard A. C., Rabies virus vaccines: Is there a need for a pan-lyssavirus vaccine? Vaccine 30, 7447–7454 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Roche S., Rey F. A., Gaudin Y., Bressanelli S., Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315, 843–848 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Roche S., Bressanelli S., Rey F. A., Gaudin Y., Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313, 187–191 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Doms R. W., Keller D. S., Helenius A., Balch W. E., Role for adenosine triphosphate in regulating the assembly and transport of vesicular stomatitis virus G protein trimers. J. Cell Biol. 105, 1957–1969 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Blois H., Tuffereau C., Blancou J., Artois M., Aubert A., Flamand A., Oral immunization of foxes with avirulent rabies virus mutants. Vet. Microbiol. 23, 259–266 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Lafay F., Bénéjean J., Tuffereau C., Flamand A., Coulon P., Vaccination against rabies: Construction and characterization of SAG2, a double avirulent derivative of SADBern. Vaccine 12, 317–320 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Ito N., Okamoto T., Sasaki M., Miyamoto S., Takahashi T., Izumi F., Inukai M., Jarusombuti S., Okada K., Nakagawa K., Fujii Y., Nishiyama S., Masatani T., Sawa H., Sugiyama M., Safety enhancement of a genetically modified live rabies vaccine strain by introducing an attenuating Leu residue at position 333 in the glycoprotein. Vaccine 39, 3777–3784 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Schmidt C., Grünberg J., Kruppa J., Formation of heterotrimers between the membrane-integrated and the soluble glycoproteins of vesicular stomatitis virus leads to their intracellular cotransport. J. Virol. 66, 2792–2797 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morimoto K., Iwatani Y., Kawai A., Shedding of Gs protein (a soluble form of the viral glycoprotein) by the rabies virus-infected BHK-21 cells. Virology 195, 541–549 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Dietzschold B., Wiktor T. J., Wunner W. H., Varrichio A., Chemical and immunological analysis of the rabies soluble glycoprotein. Virology 124, 330–337 (1983). [DOI] [PubMed] [Google Scholar]
- 29.Dolnik O., Volchkova V., Garten W., Carbonnelle C., Becker S., Kahnt J., Ströher U., Klenk H.-D., Volchkov V., Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 23, 2175–2184 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bukreyev A., Yang L., Fricke J., Cheng L., Ward J. M., Murphy B. R., Collins P. L., The secreted form of respiratory syncytial virus G glycoprotein helps the virus evade antibody-mediated restriction of replication by acting as an antigen decoy and through effects on Fc receptor-bearing leukocytes. J. Virol. 82, 12191–12204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ressing M. E., van Leeuwen D., Verreck F. A. W., Keating S., Gomez R., Franken K. L. M. C., Ottenhoff T. H. M., Spriggs M., Schumacher T. N., Hutt-Fletcher L. M., Rowe M., Wiertz E. J. H. J., Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion. J. Virol. 79, 841–852 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hueffer K., Khatri S., Rideout S., Harris M. B., Papke R. L., Stokes C., Schulte M. K., Rabies virus modifies host behaviour through a snake-toxin like region of its glycoprotein that inhibits neurotransmitter receptors in the CNS. Sci. Rep. 7, 12818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belot L., Ouldali M., Roche S., Legrand P., Gaudin Y., Albertini A. A., Crystal structure of Mokola virus glycoprotein in its post-fusion conformation. PLOS Pathog. 16, e1008383 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng W. M., Fedosyuk S., English S., Augusto G., Berg A., Thorley L., Segireddy R. R., Bowden T. A., Douglas A. D., Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies. bioRxiv 2022.02.28.482293 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.C. E. Rupprecht, A. R. Fooks, B. Abela-Ridder, Laboratory Techniques In Rabies (World Health Organization, 2019), pp. 2; https://apps.who.int/iris/handle/10665/310837.
- 36.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A. J., Berger B., Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liebschner D., Afonine P. V., Baker M. L., Bunkóczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using x-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr D Struct Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirdita M., Ovchinnikov S., Steinegger M., ColabFold - Making protein folding accessible to all. bioRxiv 10.1101/2021.08.15.456425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Melo G. D., Sonthonnax F., Lepousez G., Jouvion G., Minola A., Zatta F., Larrous F., Kergoat L., Mazo C., Moigneu C., Aiello R., Salomoni A., Brisebard E., De Benedictis P., Corti D., Bourhy H., A combination of two human monoclonal antibodies cures symptomatic rabies. EMBO Mol. Med. 12, e12628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.F. Mareuil, O. Doppelt-Azeroual, H. Ménager, A public Galaxy platform at Pasteur used as an execution engine for web services (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1
Movie S1