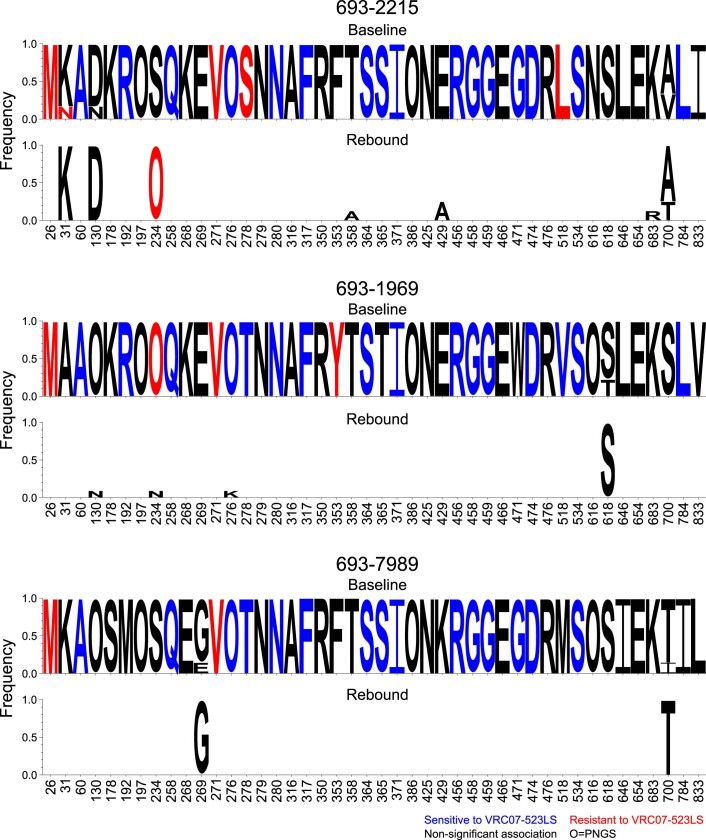
Extended Data Fig. 3. Mutations at VRC07-523LS signature sites.
All HXB2 sites significantly associated with sensitivity/resistance to VRC07-523LS from Bricault et al.1 are shown. At these sites, amino acids are shown as logos with height of the letter indicating its frequency. O represents Asn in an N-linked glycan sequon (N-X-S/T where X is not Pro). For each participant, top logos show the baseline Envs, and bottom show rebound Envs. To highlight differences, amino acids in the rebound Envs that are invariant from the baseline Envs are whitened out. If the baseline Envs had two or more variants, from which one was found in the rebound Envs, those variants are still shown (for example site 31 in participant 693-2215). Blue amino acids are associated with sensitivity to VRC07-523LS, red with resistance, and black showed no significant associations.