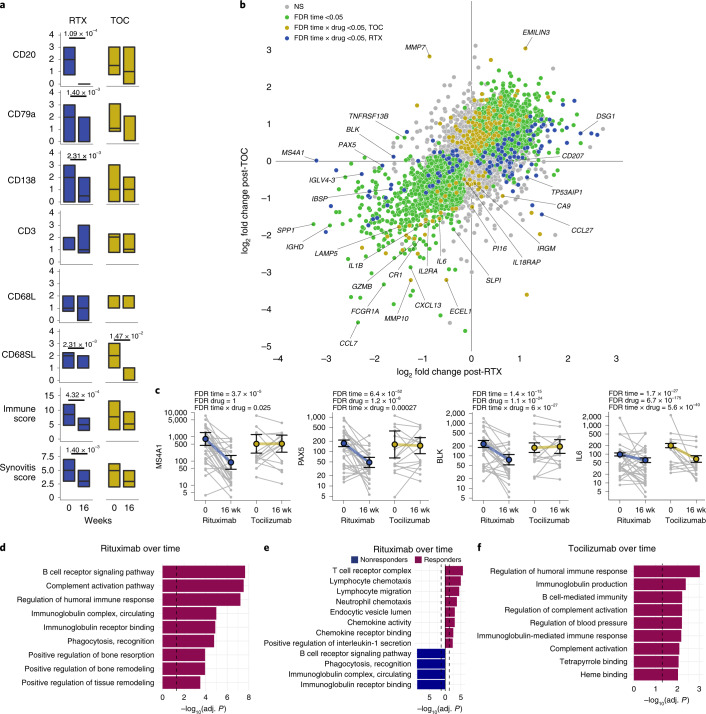
Fig. 5. Histological and molecular analysis of paired pre- and post-treatment synovial biopsies.
a, Semiquantitative histological scores of synovial immune cells at baseline and 16 weeks in patients treated with rituximab and tocilizumab. Boxplots showing median and first and third quartiles. P values shown when <0.05, two-sided Wilcoxon signed-rank test (paired) comparing baseline and 16 weeks, adjusted for multiple testing by FDR; n = 65 patients with matched baseline and 16-week samples (41 randomized to rituximab, 24 to tocilizumab). b, Scatter plots comparing longitudinal gene expression changes between drugs over 16 weeks of treatment in 88 paired biopsies from 44 patients following treatment with rituximab (n = 29) or tocilizumab (n = 15). log2 fold change in expression following rituximab or tocilizumab is represented on the x and y axis, respectively. Genes equally affected by each drug lie along the line of identity. Fold change and statistical analysis of longitudinal differential gene expression were calculated by negative binomial general linear mixed-effects model. Genes in green show significant (FDR < 0.05) overall change in expression over time; those in blue/yellow show significantly differential change in expression over time between the two drugs based on significant (FDR < 0.05) interaction term time × medication (Methods). Genes with greater absolute fold change following rituximab or tocilizumab are shown in blue and yellow, respectively. c, Scatter plots for selected genes with colored points showing regression line of fitted mixed-effects model, with error bars showing 95% CIs (fixed effects). Gray points and lines show raw paired count data, with numbers as per the analysis above. d–f, Pathway analysis using a two-sided hypergeometric test to enrich downregulated genes between baseline and 16 weeks in patients treated with rituximab (d), responders and nonresponders to rituximab (e) and responders to tocilizumab (f). Dashed line indicates adjusted P = 0.05 (Bonferroni adjustment).