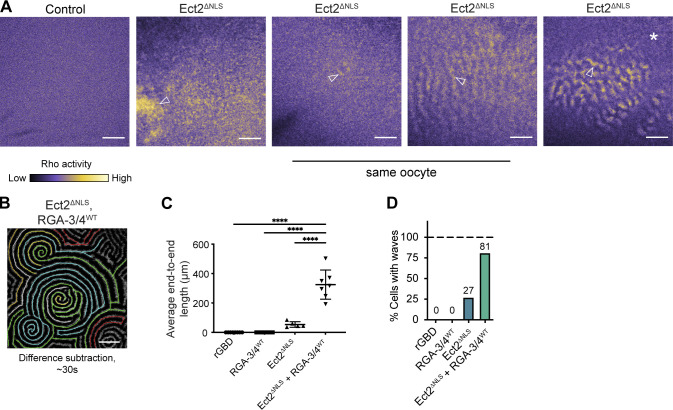
Figure S3.
Variation in Rho activity patterns in oocytes expressing Ect2 alone and quantification for oocytes coexpressing Ect2 and RGA. (A) All oocytes express probe for active Rho (GFP-rGBD). Panel 1, rGBD only; panels 2–5, examples of phenotypes from Ect2ΔNLS overexpression: static patches of Rho activity but no traveling waves (panel 2, arrowhead); tiny cluster of waves (panel 3, arrowhead) and diffuse wave patterns (panel 4, arrowhead) in same oocyte; wave patches (panel 5, arrowhead), surrounded by dormant cortex (panel 5, asterisk); scale bars = 50 µm. (B) Example still-frame difference subtraction of oocyte from (Fig. 4 C), showing segmentation process for measuring end-to-end lengths of cortical waves; scale bar = 50 µm. (C) One-way ANOVA with Tukey post hoc test for multiple comparisons, comparing end-to-end lengths across experimental groups. Each dot represents a single oocyte; group mean ± SD; data distribution was assumed to be normal but was not formally tested. Cells coexpressing Ect2ΔNLS and RGA-3/4WT are significantly different from all other groups; controls, n = 8; RGA-3/4WT, n = 7; Ect2ΔNLS, n = 11; Ect2ΔNLS + RGA-3/4WT, n = 12; seven experiments; ****, P < 0.0001. (D) Plot of percentage of cells displaying cortical waves across each experimental condition; controls, n = 8; RGA-3/4WT, n = 7; Ect2ΔNLS, n = 36; Ect2ΔNLS + RGA-3/4WT, n = 37; 13 experiments.