Figure 5.
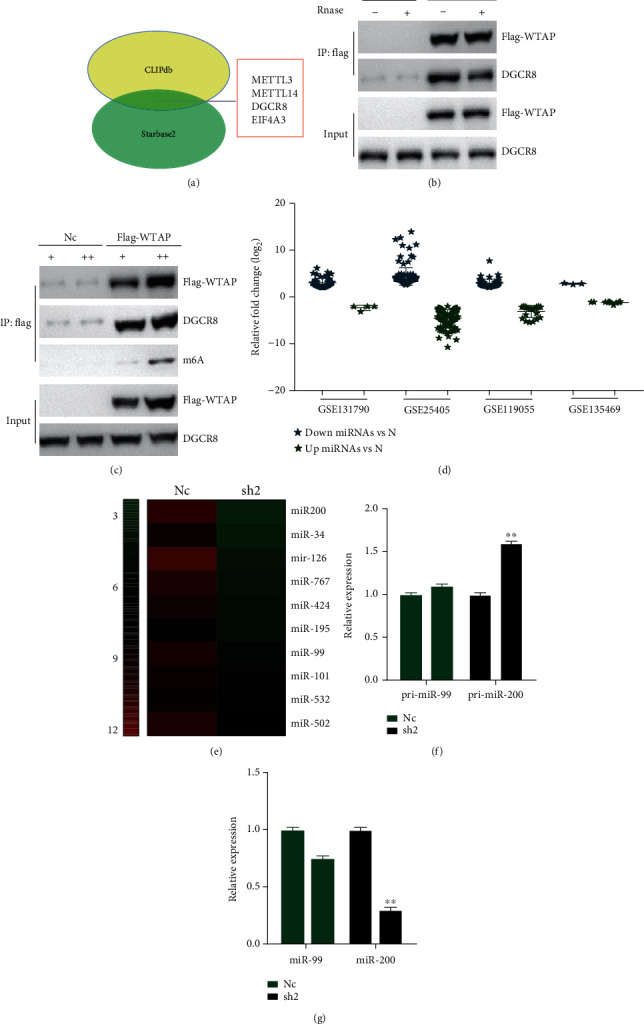
WTAP-dependent m6A demethylation regulates the processing of miR-200 via DGCR8. (a) CLIPdb and starBase2 databases were used to predict the interaction proteins of WTAP. (B) Coimmunoprecipitation of the WTAP-interacting protein DGCR8. A2780 cells were crosslinked before the immunoprecipitation. WTAP gene was tagged by flag in advance. Ribonuclease was applied as indicated. Immunoglobulin G antibody was used as negative control for the experiment. (c) Immunoprecipitation of DGCR8, WTAP, and associated RNA from control cells or cells overexpressing WTAP (+ stands for control, ++ represents over expressed WTAP flag). Western blot and immunoblot experiments were done using the antibodies as depicted. (d) Downregulated and upregulated miRNAs were analyzed and selected in OC tissues compared to adjacent normal tissues from Gene Expression Omnibus microarray profiles (GSE131790, GSE25405, GSE119055, and GSE135469). (e) Gene-wide expression profiling chip was fabricated in A2780-Nc and A2780-sh2 cells to screen differentially expressed miRNAs after WTAP knockdown. (f) Pri-miR-99 and pri-miR-200 were detected by RT-qPCR upon WTAP silencing in A2780 cells. (g) MiR-99 and miR-200 expressions were detected by RT-qPCR upon WTAP silencing in A2780 cells. Annotation: RNase = ribonuclease; IP = immunoprecipitation; IgG = immunoglobulin G; oe = overexpressing).
