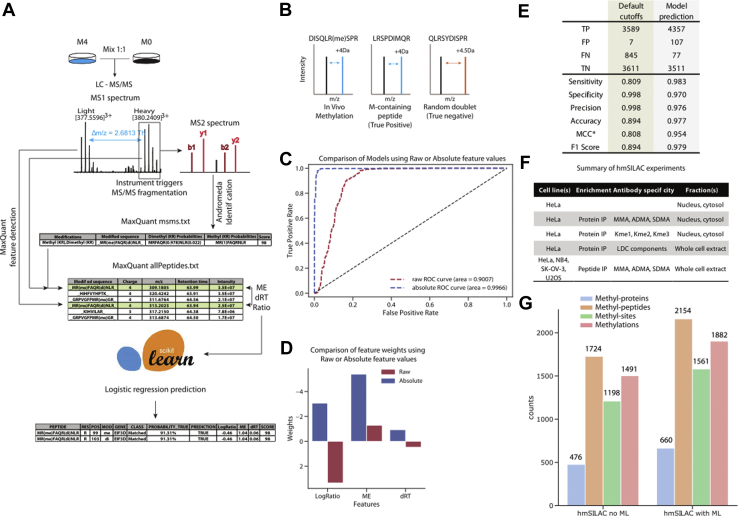
Fig. 1.
Rationale of hmSEEKER, development of the machine learning (ML) model and analysis of hmSILAC data.A, schematic representation of the hmSEEKER workflow upon metabolic labeling with stable isotope–encoded methionine (M). Cells grown in “light” (M0) and “heavy” (M4) media are mixed in 1:1 proportion, and then proteins are extracted, digested, and analyzed by LC–MS/MS. MS spectra are analyzed with MaxQuant to obtain peptide and PTM identifications. The hmSEEKER software reads MaxQuant peptide identifications and, for each methyl-peptide that passes the quality filters, finds its corresponding MS1 peak, then searches the corresponding heavy/light counterpart. A peak doublet is defined by the difference in the retention time (dRT), the log-transformed intensity ratio (LogRatio), and the deviation between expected and observed m/z delta (mass error [ME]); these parameters are used to predict if the peak pair is a true hmSILAC doublet or a false positive. B, schematic examples of true positive and true negative doublets. C, receiving operator characteristic (ROC) curves obtained by testing the models trained by using either the raw or absolute value of the features. D, representation of the ML model weights with (blue) and without (red) taking the absolute values of the features. E, comparison of the performance of hmSEEKER before and after the introduction of the ML predictor (∗MCC = Matthew's correlation coefficient). F, summary of hmSILAC experiments analyzed to produce the orthogonally validated methyl-proteome. A detailed description of these experiments is available in supplemental Table S1. G, composition of the high-confidence hmSILAC R-methyl-proteome before (left) and after (right) the implementation of the ML model. hmSILAC, heavy methyl stable isotope labeling with amino acids in cell culture; MS, mass spectrometry; PTM, post-translational modification.