Figure 9.
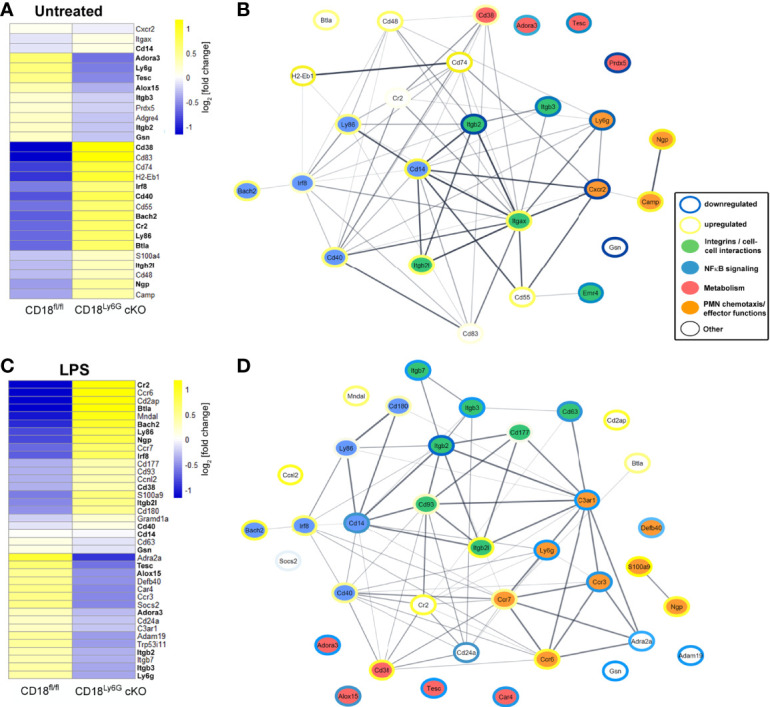
Transcriptomes and functional interaction networks of PMN-associated genes directly after isolation or upon over-night treatment with LPS. PMN were sorted from CD18fl/fl and CD18Ly6G cKO mice (each n=3) and RNA-seq was performed from untreated PMN (A) or LPS-treated PMN (C). Expression of indicated PMN-associated genes was analyzed using CLC Genomics Workbench. Genes being differentially regulated both in LPS-treated and freshly isolated PMN are shown in bold (A, C). Predicted interaction networks of the encoded proteins were being visualized using the STRING package in Cytoscape. Genes shown in the interaction networks of untreated PMN (B) or LPS-treated PMN (D) were categorized into 4 groups affecting either PMN cell-cell interactions, NFκB singaling, PMN metabolism or PMN chemotaxis and PMN effector functions. Colored borders illustrate the degree of the up- or downregulation (log fold change) found for the genes of PMN isolated from CD18Ly6G cKO mice as compared to PMN isolated from CD18fl/fl mice. Legend in (B, D).
