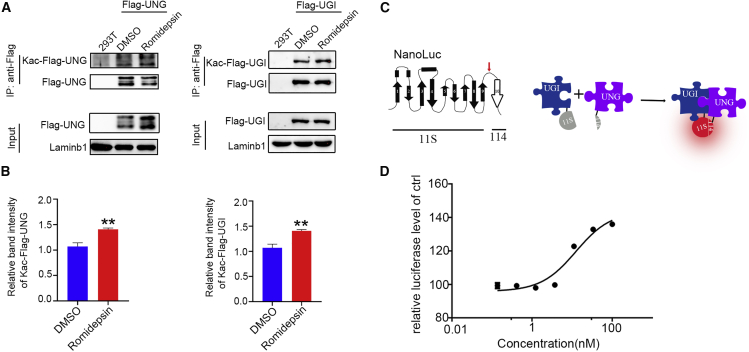
Figure 7.
HDAC inhibition increased the acetylation level of UNG and UGI and enhanced their interaction
(A) Western blotting (WB) showing the lysine acetylation level of UNG and UGI. WB analysis for the co-immunoprecipitation (co-IP) of whole-cell proteins from HEK293T control cells or cells transfected with empty or FLAG-UNG plasmids (top panels). WB analysis for input whole-cell proteins (bottom panels). The transfected cells received DMSO or romidepsin treatment as indicated. (B) Quantification of the relative acetylation level of UNG and UGI of cells treated with romidepsin to that of DMSO-treated cells. (C) Schematic diagram showing the NanoBiT assay. NanoLuc topology model 9 consisting of a 10-stranded beta-barrel (left panel). The split point (red arrow) occurs between residues 156th and 157th amino acids, generating a large fragment of 156 amino acids, referred to as 11S, and a small fragment of 11 amino acids, referred to as 114. The interaction of fusion proteins pulled 11S and 114 together to form a functional luciferase (right panel). (D) Dose-dependent enhancement of romidepsin to the interaction between UNG and UGI. Data are represented as mean ± SD; asterisks indicate statistically significant differences between DMSO-treated cells and HDAC inhibitor-treated cells (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).