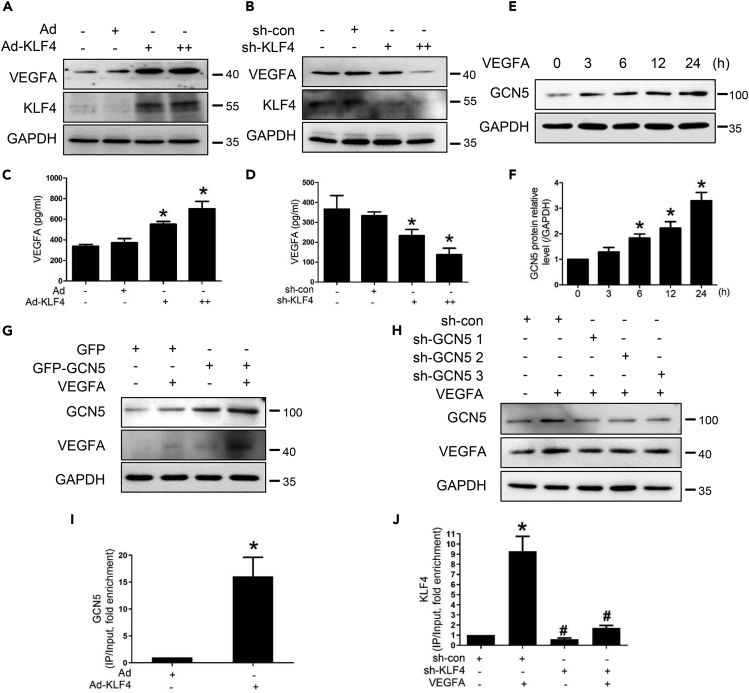
Figure 4.
GCN5 participates in the feedback regulatory loop between KLF4 and VEGFA
(A–B) After HEMECs were infected with Ad-KLF4 or sh-KLF4 adenovirus for 36 h, VEGFA and KLF4 protein levels were detected by Western blotting.
(C–D) We treated HEMECs with Ad-KLF4 or sh-KLF4 adenovirus for 36 h and detected VEGFA levels in cell culture supernatant by ELISA. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. Ad or sh-con, #p < 0.05 vs. VEGFA; one-way ANOVA.
(E) HEMECs were stimulated with VEGFA for different time intervals, and GCN5 expression level was assessed by Western blotting.
(F) Densitometric analyses for Western blots in (E). Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 h, one-way ANOVA.
(G–H) HEMECs were transfected with expression plasmids for GCN5 or three GCN5-specific knockdown plasmids (sh-GCN5 1, sh-GCN5 2, and sh-GCN5 3) and treated with VEGFA. Western blotting analysis was performed for GCN5 and VEGFA expressions.
(I) ChIP analysis of GCN5 occupancy at the VEGFA promoter in HEMECs that were infected with Ad or Ad-KLF4. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. Ad, Student’s t-test.
(J) ChIP analysis of KLF4 occupancy at the VEGFA promoter in HEMECs that were infected with sh-con or sh-KLF4 for 24 h and then treated with VEGFA for an additional 12 h. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. sh-con, #p < 0.05 vs. VEGFA, one-way ANOVA.