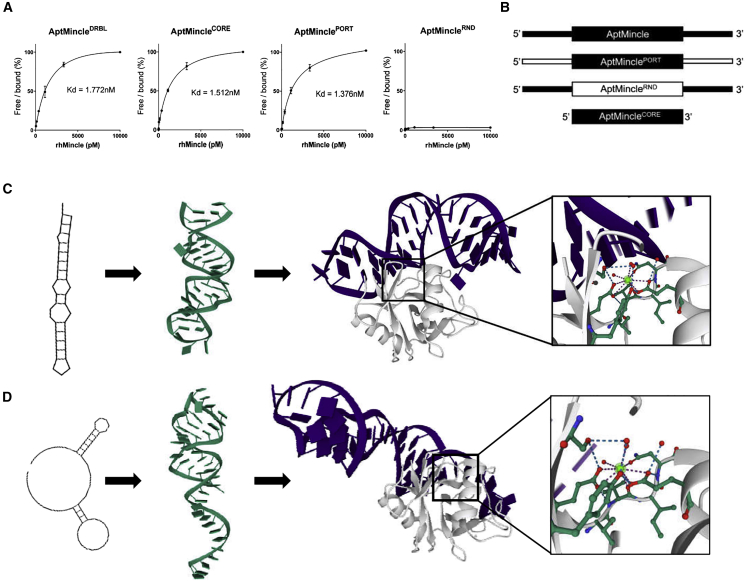
Figure 1.
Characterization of the in vitro function of aptamers with Mincle affinity
(A) Binding characteristics of AptMincle, and its modified counterparts, with rhMincle as determined by ELONA. (B) Graphical depiction of the modifications made to the aptamer sequences (black, original; white, modified). (C) The predicted secondary and tertiary structures and docking simulation of AptMincleCORE (purple) with Mincle (white). Box: highlighted predicted region of interaction surrounding a calcium molecule. (D) The predicted secondary and tertiary structures and docking simulation of AptMincleRND (purple) with Mincle (white). Box: highlighted predicted region of interaction surrounding a calcium molecule. Dissociation constants were calculated on GraphPad Prism 8 using a non-linear regression binding analysis with assumed one-site target parameters.