Abstract
Patient: Male, 57-year-old
Final Diagnosis: Gastric cancer
Symptoms: Non
Medication:—
Clinical Procedure: —
Specialty: Oncology
Objective:
Challenging differential diagnosis
Background:
Clonal hematopoiesis is the production of a specific single clonal type of cell in the blood and is often found in cancer genomic profiling tests. When the clone carries a pathogenic variant, it may be important to differentiate between somatic or germline origin. The variant in the blood that has a lower minor allele frequency could reflect heterozygous germline origin, somatic mosaicism, and clonal hematopoiesis. It is important to evaluate suspected variants to determine the course of treatment and follow-up of the patient, depending on the patient’s medical condition and family situation.
Case Report:
We report a 57-year-old Japanese man with gastric cancer who underwent a cancer genomic profiling test searching for therapeutic agents. The profiling test detected a variant, TP53 c.559+2T>G minor allele frequencies of 9% (168/1865) in tumor tissue and 29.1% (58/199) in paired blood. Since the TP53 variant has the possibility of Li-Fraumeni syndrome, ancillary testing was performed using fingernails, buccal swab, and blood specimens. The genomic analysis revealed no TP53 variant in his fingernails. The patient had previously received platinum-based chemotherapies, suggesting that the variant reflected treatment-induced clonal hematopoiesis.
Conclusions:
Identifying clonal hematopoiesis when performing genomic profiling tests for patients with cancer is important. Examining multiple tissues to determine whether a variant arises from clonal hematopoiesis or is of germline origin can provide more accurate genetic information and improve patient follow-up care.
Keywords: Clonal Hematopoiesis, Li-Fraumeni Syndrome, Tumor Suppressor Protein p53
Background
The increasing use of next-generation sequencing has enabled the development of cancer treatment strategies that target certain gene profiles using effective therapeutic agents. More than 300 cancer-associated genes have been identified as actionable variants [1]. Tests of tumor tissue and paired blood show that the percentage of variants in the latter is approximately 50% in general; these are usually assumed to be of germline origin. Variants in the blood that have a lower minor allele frequency (MAF) could reflect post-zygotic mosaicism [2].
Additionally, the possibility of clonal hematopoiesis (CH), which is a skewing in blood cell clones, should be considered if the patient is undergoing cancer treatment [3]. Whether variants are heterozygous germline, somatic mosaicism, or CH will determine the patient’s treatment and/or follow-up strategy; therefore, evaluating the suspected variant is important, although the necessity of doing so varies depending on the patient’s medical condition and family situation.
In this report, we describe a patient with gastric cancer who underwent cancer genomic profile testing of both the tumor and paired blood, whereupon TP53 pathogenic variants were detected in both specimens. Additional ancillary tissue testing suggested the possibility of CH. We discuss the clinical implications of our results as well as genetic counseling.
Case Report
A Japanese man was diagnosed with stage IV gastric cancer with distant lymph node metastasis as his first cancer at age of 53; the pathological diagnosis was consistent with poorly-differentiated adenocarcinoma. His initial chemotherapy regimen was 8 cycles of tegafur/gimeracil/oteracil (S-1) plus cisplatin, which was followed by 3 cycles of S-1, 7 cycles of nanoparticle albumin-bound-paclitaxel plus ramucirumab, and nivolumab. After the cancer progressed, he most recently received irinotecan, trifluridine plus tipiracil hydrochloride, and capecitabine plus oxaliplatin.
At age 57 years (3 years 4 months after his initial diagnosis with gastric cancer), the patient underwent a gastric biopsy; the specimen and paired blood were subjected to a tumor genome profiling test (NCC-OncoPanel System, Sysmex Corporation, Kobe, Japan). He was found to have a variant, NM_000546.5(TP53): c.559+2T>G, with a MAF of 9% (168/1865) in the tumor and 29.1% (58/199) in the paired blood. The splice donor variant of intron 5 lead to a loss of function at the authentic splice site; this variant was deemed to be “pathogenic” according to the American College of Medical Genetics guidelines. The MAFs of the other 4 pathogenic variants detected in tumor specimens ranged from 5.2% to 52.1%, which were within the average range reported previously [4] (Table 1).
Table 1.
The MAF values are provided for each variant. The TP53: c.559+2T>G variant was present in the tumor and paired blood samples. The remaining variants were not present in the paired blood.
| cDNA | Protein | MAF tumor (total reads) | MAF blood (total reads) | |
|---|---|---|---|---|
| ALK * | c.938A>C | p.(Lys313Thr) | 5.2% (122/2338) | – |
| CDKN2A ** | c.248A>G | p.(His83Arg) | 40.7% (767/1884) | – |
| CREBBP *** | c.5954G>A | p.(Arg1985His) | 11.2% (35/313) | – |
| TP53 **** | c.578A>G | p.(His193Arg) | 52.1% (613/1177) | – |
| TP53 | c.559+2T>G | – | 9% (168/1865) | 29.1% (58/199) |
ALK transcript NM_004304.5;
CDKN2A transcript NM_000077.5;
CREBBP transcript NM_004380.3;
TP53 transcript NM_000546.5.
MAF – minor allele frequency.
The patient did not meet the Chompret or classical criteria for Li-Fraumeni syndrome (LFS), even though gastric cancer has been associated with LFS [5,6]. The low MAF suggested the possibility of post-zygotic mosaicism; 7 of the patient’s first- and second-degree family members had cancer, but mostly when in their 70s and 80s. Moreover, his 3 offspring (aged 30, 27, and 24 years) did not have any malignancies. Hematological malignancies such as myelodysplastic syndromes and Waldenström macroglobulinemia were ruled out because his laboratory findings (including complete blood counts) were within the normal range. His history of having received platinum-based agents such as cisplatin and oxaliplatin suggested chemotherapy-induced CH. However, to clarify whether the variant was CH, a validation test using different specimens seemed to be necessary. Although another tumor specimen (biological replicate) was not available, we offered him to have a confirmation test using nail, oral swabs, and another blood specimens, considering the minimum invasiveness. Informed consent was obtained from the patient to perform molecular DNA analysis, which was also approved by the Ethics Review Board for clinical studies at the National Hospital Organization Nagoya Medical Center (No. 2014-756).
Four milliliters of peripheral blood were drawn into an EDTA-containing tube. The buccal mucosa cells were collected using Sterile DNA-Free Large Round Foam Swab. Fingernails were cut with clean clippers and collected. The genomic DNAs were extracted from the blood using the Gentra Puregine Blood Kit (Qiagen, Inc., Valencia, CA), the buccal swab using the QIAamp DNA Mini Kit (Qiagen, Inc., Valencia, CA), and fingernail using ISOHAIR (Nippon Gene, Inc., Tokyo, Japan), according to the manufacturer’s protocols, respectively. PCR direct sequencing was performed on exon 5 and its exon-intron boundaries because the variant (c.559+2T>G) was at the 5’ splice site beginning intron 5 of TP53 gene. Each PCR reaction contained 1x PrimeSTAR GXL buffer, 200 uM each dNTP mix, 0.5U DNA polymerase, 0.2 uM each PCR primer, 25 ng template DNA purified from each sample, and ddH2O to give a total volume of 20 μl. The PCR condition was 40 cycles of 98°C for 10 s, 60°C for 15 s, and 68°C for 1 min. The PCR primers were designed as follows: forward primer 5’-ACACGCAAATTTCCTTCCAC-3’ and reverse primer 5’-AGTACTCCCCTGCCCTCAAC-3’. The amplified products were sequenced using an Applied Biosystem 3500 automated sequencing system (Applied Biosystems, Waltham, MA).
The TP53 variant (c.559+2T>G) was identified in the patient’s blood and buccal swab, but not in the fingernails (Figure 1). Pathological examination of the tumor tissue revealed that it was infiltrated by hematopoietic cells such as lymphocytes and histiocytes. The percentage of lymphocytes in the tumor specimen was approximately 50% (Figure 2), suggesting that the TP53 pathological variant in the tumor tissue originated from hematopoietic cells.
Figure 1.
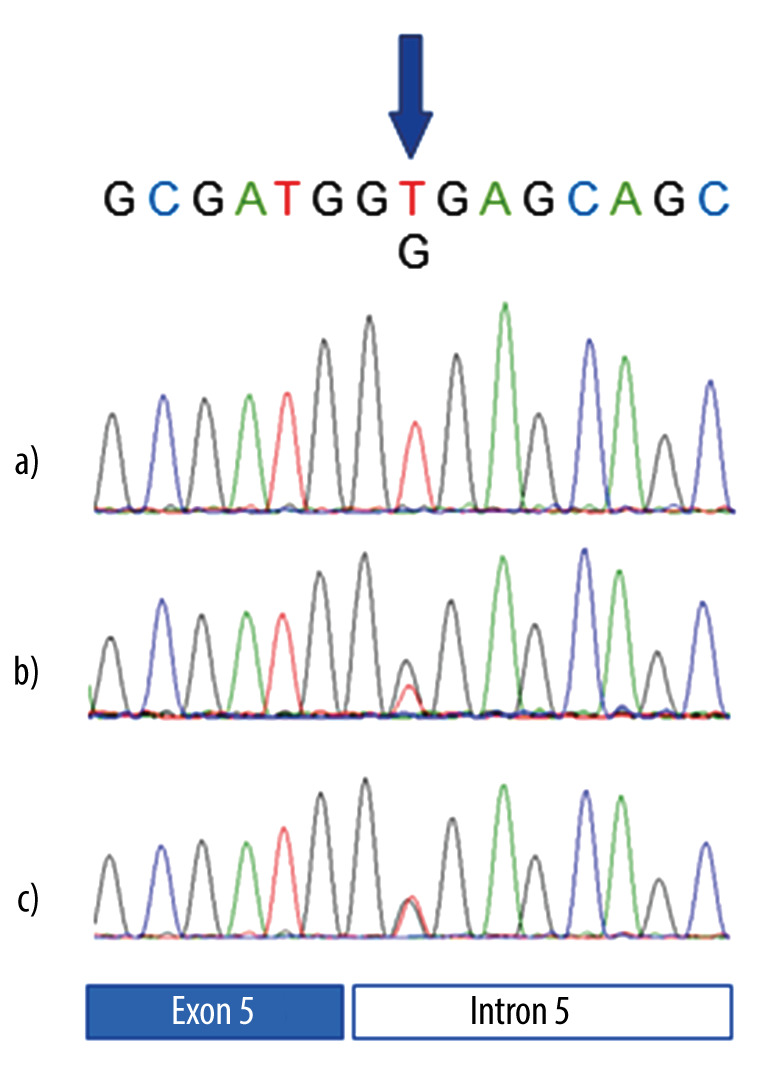
Sequencing analysis of the TP53 gene in the patient. a), b), and c) represent fingernails, blood, and buccal swab, respectively. The pathogenic variant was detected in the blood and the buccal swab, but not in the fingernail.
Figure 2.
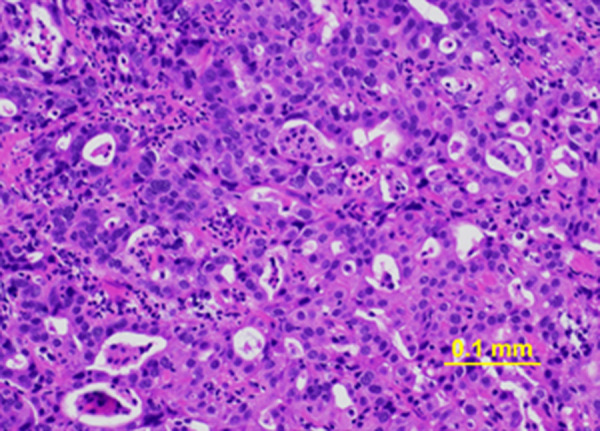
Hematoxylin and eosin-stained gastric cancer biopsy sample. The estimated proportions of tumor cells, inflammatory infiltrates, and normal cells are 30–40%, 50%, and 10%, respectively.
Taken together, we assumed the variant did not indicate post-zygotic mosaicism but rather CH. The patient and 2 of his off-spring attended a genetic counseling session, in which we explained that the variant was considered as CH, unlikely to be of germline origin. The patient and his children accepted the test results and were satisfied with the explanation. The children did not wish to have genetic testing because of the additional cost and because none of them had developed cancer. We also recommended that he undergo regular blood tests for monitoring, as CH can occasionally develop into hematological malignancies such as myelodysplastic syndromes.
Discussion
Although blood DNA is thought to represent the germline, genetic cancer risk assessment after cancer profiling testing should be performed with caution, especially when assessing variants in TP53 given their association with LFS. Approximately 20% of TP53 variants identified by multigene and single-gene tests are thought to be acquired aberrant clonal expansions, mostly caused by CH [7]. Almost all patients with CH in Weber-Lassalle et al’s study showed MAFs of less than 20%; however, several were between 25% and 30% [7]. Our patient showed a relatively high MAF (29.1%); however, he had only subtle evidence of LFS in his family history. His previous intensive treatment with DNA-damaging agents necessitated that CH be considered; hence, we performed ancillary tissue tests on non-blood samples and presumed that they were negative for suspected germline mutations.
The TP53 variant was detected in the tumor tissue owing to the presence of infiltrated blood cells. In several studies of patients with CH, TP53 variants were not detected in tumors [8–10]. A small fraction of the variant may have been detected in our patient given that intratumoral heterogeneity has previously been shown [11]. If the variant had originated from post-zygotic mosaicism and was associated with tumorigenesis, the MAF value would be expected to be higher in the tumor specimens, as previously reported [12]. Although we were not able to completely rule out post-zygotic mosaicism, we concluded the patient likely had CH, based on his family and chemotherapy histories. The CH variant detected in the buccal swab was presumed to be due to contaminated blood cells in the saliva. Analyses of several tissue types and overall assessment are needed given the lack of established methods to differentiate CH from post-zygotic mosaicism. Further analysis using another non-blood contaminated tissue such as skin fibroblast, or serial blood sampling was ideal. However, these were not available in our case, because the patient desired minimum invasiveness and died due soon thereafter due to disease progression.
Anticancer drugs have recently been reported to be a leading cause of CH. Platinum and topoisomerase II inhibitors are associated with CH mutations in DNA damage-response genes, such as TP53, PPM1D, and CHEK2 [13]. Our patient received cytotoxic chemotherapy agents, including platinum, over a 4-year period. An assessment of CH is important for patient management because this condition reportedly increases the risk of hematologic malignancies approximately 12.9 times [3]. Therefore, regular observation of blood count is recommended for clinical management.
Herein, we discussed CH as a secondary finding on cancer panel testing; however, in the near future, the assessment of TP53 CH will be more important for the identification of therapeutic targets given that clinical trials and the development of anti-tumor drugs targeting TP53 mutations are underway [14,15. When variants of TP53 are detected in tumor specimens, as in our patient, it is necessary to accurately determine whether they are truly tumor-derived or rather represent CH in contaminating blood, as this is necessary for planning the optimal treatment. Our described approach for evaluating the suspected variants can be useful in many situations.
Conclusions
Genomic profiling tests can identify CH even in non-elderly patients who have received chemotherapy. Both high MAF values and massive blood infiltration into the tumor tissue can lead to erroneous interpretations under routine specimen settings. Testing of other tissue specimens in addition to tumor and blood can lead to more accurate results. A thorough assessment of CH will be helpful for providing guidance and follow-up care for patients.
Acknowledgments
We thank the patient for agreeing to participate. This study was partly supported by a Grant-in-Aid for Scientific Research from the Japan Agency for Medical Research and Development (AMED).
Footnotes
Declaration of Figures’ Authenticity
All figures submitted have been created by the authors who confirm that the images are original with no duplication and have not been previously published in whole or in part.
References:
- 1.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batalini F, Peacock EG, Stobie L, et al. Li-Fraumeni syndrome: not a straightforward diagnosis anymore – the interpretation of pathogenic variants of low allele frequency and the differences between germline PVs, mosaicism, and clonal hematopoiesis. Breast Cancer Res. 2019;21:1–10. doi: 10.1186/s13058-019-1193-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sunami K, Ichikawa H, Kubo T, et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 2019;110:1480–90. doi: 10.1111/cas.13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funato M, Tsunematsu Y, Yamazaki F, et al. Characteristics of Li-Fraumeni syndrome in Japan: A review study by the special committee of JSHT. Cancer Sci. 2021;112:2821–34. doi: 10.1111/cas.14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masciari S, Dewanwala A, Stoffel EM, et al. Gastric cancer in individuals with Li-Fraumeni syndrome. Genet Med. 2011;13:651–57. doi: 10.1097/GIM.0b013e31821628b6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weitzel JN, Chao EC, Nehoray B, et al. Somatic TP53 variants frequently confound germ-line testing results. Genet Med. 2018;20:809–16. doi: 10.1038/gim.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber-Lassalle K, Harter P, Hauke J, et al. Diagnosis of Li-Fraumeni syndrome: Differentiating TP53 germline mutations from clonal hematopoiesis: Results of the observational AGO-TR1 trial. Hum Mutat. 2018;39:2040–46. doi: 10.1002/humu.23653. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell RL, Kosche C, Burgess K, et al. Misdiagnosis of Li-Fraumeni syndrome in a patient with clonal hematopoiesis and a somatic TP53 mutation. J Natl Compr Cancer Netw. 2018;16:461–66. doi: 10.6004/jnccn.2017.7058. [DOI] [PubMed] [Google Scholar]
- 10.Ito M, Fujiwara Y, Kubo T, et al. Clonal hematopoiesis from next generation sequencing of plasma from a patient with lung adenocarcinoma: A case report. Front Oncol. 2020;10:113. doi: 10.3389/fonc.2020.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–68. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 12.Behjati S, Maschietto M, Williams RD, et al. A pathogenic mosaic TP53 mutation in two germ layers detected by next generation sequencing. PLoS One. 2014;9:e96531. doi: 10.1371/journal.pone.0096531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52:1219–26. doi: 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malekzadeh P, Yossef R, Cafri G, et al. Antigen experienced T cells from peripheral blood recognize p53 neoantigens. Clin Cancer Res. 2020;26:1267–76. doi: 10.1158/1078-0432.CCR-19-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seligmann JF, Fisher DJ, Brown LC, et al. Inhibition of WEE1 is effective in TP53- and RAS-mutant metastatic colorectal cancer: A randomized trial (FOCUS4-C) comparing adavosertib (AZD1775) with active monitoring. J Clin Oncol. 2021;39:3705–15. doi: 10.1200/JCO.21.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]


