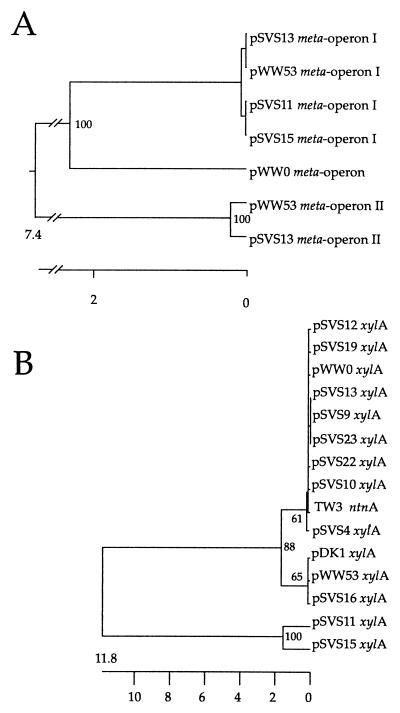
FIG. 7.
Phylogenetic comparison of selected DNAs from the different meta pathway operons and upper pathway operons of several pSVS plasmids of types A, B, and C and corresponding sequences of other TOL plasmids. (A) Alignment based on 730 nucleotides within xylL and xylT (Fig. 3). (B) Alignment based on 395 nucleotides at the 3′ end of xylA. From the pWW0 sequence of the xylLT region, 36 nucleotides (positions 4229 to 4265) were left out, apparently introduced by mistake (P. Golyshin, personal communication). Trees were constructed by using CLUSTAL V. The scales below the trees indicate the sequence distances as the number of substitutions per 100 nucleotides. Only bootstrapping values above 50% occurring in 100 replicate trees, obtained by using the PARSIMONY program, are shown in the nodes.