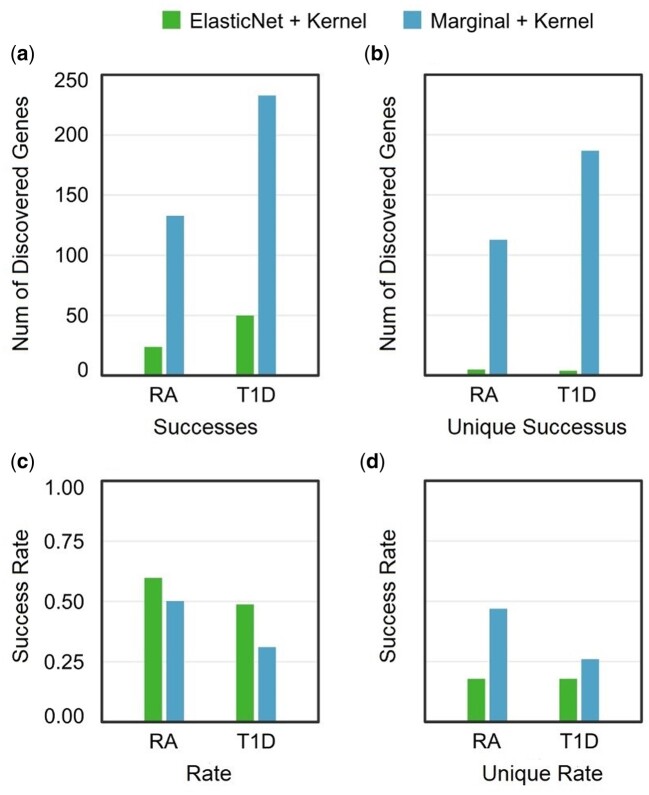
Figure 5.
Comparison of marginal effect- and ElasticNet-based feature selection in WTCCC data. The two disentangled protocols ElasticNet + Kernel (left) and Marginal + Kernel (right) are compared on two WTCCC diseases (T1D and RA). Both protocols use kernel-based feature aggregation, but have different feature selection methods. (A) Number of discovered genes (successes) which are reported as disease-associated in DisGeNET. (B) Number of discovered genes (successes) discovered exclusively by one of the two protocols under comparison. (C) Proportion (success rate) of all discovered genes which are validated by DisGeNET. (D) Proportion (success rate) of genes discovered exclusively by each protocol which are validated by DisGeNET.