Abstract
Cellular hypoxia occurs when the demand for sufficient molecular oxygen needed to produce the levels of ATP required to perform physiological functions exceeds the vascular supply, thereby leading to a state of oxygen depletion with the associated risk of bioenergetic crisis. To protect against the threat of hypoxia, eukaryotic cells have evolved the capacity to elicit oxygen-sensitive adaptive transcriptional responses driven primarily (although not exclusively) by the hypoxia-inducible factor (HIF) pathway. In addition to the canonical regulation of HIF by oxygen-dependent hydroxylases, multiple other input signals, including gasotransmitters, non-coding RNAs, histone modifiers and post-translational modifications, modulate the nature of the HIF response in discreet cell types and contexts. Activation of HIF induces various effector pathways that mitigate the effects of hypoxia, including metabolic reprogramming and the production of erythropoietin. Drugs that target the HIF pathway to induce erythropoietin production are now approved for the treatment of chronic kidney disease-related anaemia. However, HIF-dependent changes in cell metabolism also have profound implications for functional responses in innate and adaptive immune cells, and thereby heavily influence immunity and the inflammatory response. Preclinical studies indicate a potential use of HIF therapeutics to treat inflammatory diseases, such as inflammatory bowel disease. Understanding the links between HIF, cellular metabolism and immunity is key to unlocking the full therapeutic potential of drugs that target the HIF pathway.
Subject terms: Preclinical research, Inflammation
Hypoxia-dependent changes in cellular metabolism have important implications for the effective functioning of multiple immune cell subtypes. This Review describes the inputs that shape the hypoxic response in individual cell types and contexts, and the implications of this response for cellular metabolism and associated alterations in immune cell function.
Key points
Hypoxia is a common feature of particular microenvironments and at sites of immunity and inflammation, resulting in increased activity of the hypoxia-inducible factor (HIF).
In addition to hypoxia, multiple inputs modulate the activity of the HIF pathway, allowing nuanced downstream responses in discreet cell types and contexts.
HIF-dependent changes in cellular metabolism mitigate the effects of hypoxia and ensure that energy needs are met under conditions in which oxidative phosphorylation is reduced.
HIF-dependent changes in metabolism also profoundly affect the phenotype and function of immune cells.
The immunometabolic effects of HIF have important implications for targeting the HIF pathway in inflammatory disease.
Introduction
The Earth’s outer crust is coated with a thin layer of air upon which most animals depend for the provision of sufficient levels of molecular oxygen to fuel their metabolism and survival1. This delicate sheath comprises a gradient of oxygen partial pressure (pO2) that ranges from 159 mmHg at sea level to 52.8 mmHg at the summit of mount Everest — the highest point on earth at 8,848 m (ref.2). Notably, the oxygen levels in the Earth’s atmosphere have fluctuated over the course of geological time and continue to do so1. Most multicellular animals (metazoans) that inhabit the earth’s terrestrial and oceanic surfaces have evolved aerobic respiration as a metabolic strategy. This process breaks down glucose and uses oxygen as a final electron acceptor in the electron transport chain (ETC); the efficient extraction of energy from this step produces the primary cellular fuel, ATP, at levels sufficient to fuel cellular processes3. An important payoff for the evolution of this highly-efficient oxygen-dependent cellular metabolic strategy is the absolute dependence of most metazoan cells on a constant supply of oxygen in order to simply maintain bioenergetic homeostasis and survival on a minute-to-minute basis. However, exposure to conditions of hypoxia is a metabolic challenge that is common to a wide range of physiological and pathophysiological states4. For this reason, metazoan cells evolved oxygen-dependent signalling in tandem with the evolution of the capacity for oxidative metabolism, such as that involving hypoxia-inducible factor (HIF), to elicit metabolic and other adaptations to oxygen deprivation.
A growing body of evidence demonstrates that such hypoxia-dependent changes in cellular metabolism have profound implications for the effective functioning of immune cells. In this Review, we discuss the canonical and non-canonical inputs that shape the hypoxic response in individual immune cell types and contexts. We describe the implications of this response for cellular metabolism and associated alterations in immune cell function. Finally, we discuss the regulation of immunity and inflammation in the context of the therapeutic application of drugs that target hypoxia-sensitive pathways.
Physiological hypoxia
Hypoxia is frequently encountered in physiological situations, including fetal development and physical exertion5. pO2 gradients exist around every blood vessel, decreasing with increasing distance from the vessel2. During fetal development, tissues outgrow their local vascular supply, leading to regional hypoxia6. In addition, multiple tissues in the healthy adult body, including the kidney and the mucosal surface of the gastrointestinal tract, experience steep gradients of oxygen relative to other tissues as a consequence of high levels of oxygen consumption and/or limited perfusion. Importantly, the oxygen gradients in the kidney are relatively stable7–9. In skeletal muscle, higher contractile activity during exercise increases oxygen consumption, which can also lead to physiological hypoxia10. Finally, exposure to lower atmospheric pO2 caused by ascent to high altitude can lead to decreased blood oxygen levels (hypoxaemia) and systemic hypoxia11. Thus, exposure to hypoxia and its associated threats to bioenergetic homeostasis is a frequent event associated with a range of common physiological processes; however, the enhanced expression of a large number of specific genes — particularly those regulated by HIF — ensure that metabolic homeostasis is maintained.
Pathophysiological hypoxia
Tissue hypoxia is also associated with a number of pathological states and influences the activation of cellular signals, which in turn can contribute to the development and/or progression of disease12. For example, tumours that outgrow the existing local blood supply frequently develop regions of profound hypoxia, which activates a number of effector processes that promote tumour growth13. Chronic inflammation, such as that associated with rheumatoid arthritis, inflammatory bowel disease or in the context of inflammatory kidney disease can also induce tissue hypoxia by causing damage to blood vessels. This hypoxia also occurs at least in part as a consequence of increased oxygen consumption by immune cells, such as activated neutrophils, following their recruitment to the site of inflammation5. Pathophysiological hypoxia is a key feature of virtually all ischaemic diseases, including myocardial infarction, stroke and ischaemic kidney injury, and is the primary cause of necrotic cell death in these diseases14,15. Chronic pulmonary diseases such as chronic obstructive pulmonary disease are a major cause of death driven by the generation of a hypoxic state due to dysfunctional gas exchange in the lung. Many patients with obstructive sleep apnoea syndrome experience an intermittent form of hypoxia during sleep16,17. Hypoxia may also increase susceptibility to kidney injury8 and contribute to the progression of chronic kidney disease (CKD)18,19, although controversy exists as to the causality of this association20. Finally, pulmonary viral infections such as COVID-19, which is caused by the SARS-Co-V2 virus, lead to local as well as systemic pathological hypoxia21,22. Many of these conditions have a notable immune and/or inflammatory component. Indeed, hypoxia is a common microenvironmental manifestation at sites of inflammation.
Thus, as well as being a common physiological occurrence, tissue hypoxia is frequently encountered in a range of pathophysiological states associated with immunity and inflammation. Importantly, however, although the nature of physiological hypoxia is relatively stable and moderate, pathological hypoxia is often more varied in its severity and extent within the affected tissue or area, and can therefore have adverse consequences for cells, tissues and organisms5.
Responses to hypoxia
Cells require an adequate level of oxygen to maintain bioenergetic homeostasis. However, and as outlined above, cellular hypoxia is a frequent microenvironmental feature in a range of physiological and pathological conditions. Therefore, it is logical that adaptive responses to perceived hypoxia evolved coincident with the evolution of oxidative metabolism in multicellular organisms to avoid bioenergetic crisis and prevent extensive tissue damage.
Rapid responses by chemosensors
Chemosensors are neuron-like tissues that can detect systemic hypoxia and elicit a rapid, non-transcriptional adaptive response at the organism level, for example, by stimulating the respiratory centres in the medulla oblongata of the central nervous system to increase the rate and depth of breathing. For example, acute decreases in arterial pO2 induce rapid changes in ion channel activity in the carotid body that are non-transcriptional but are dependent on mitochondria-derived signals such as reactive oxygen species (ROS) and ATP levels (although the exact nature of the oxygen-sensing mechanism remains controversial). This rapid response to systemic hypoxia, though important for physiological adaptation, has been reviewed elsewhere23,24 and is not further discussed here.
Transcriptional responses to hypoxia
In addition to the systemic, adaptive response to hypoxia elicited by chemosensors, hypoxia induces a ubiquitous and highly evolutionarily conserved pathway that activates an adaptive transcriptional response at the cellular level. This pathway was first identified in studies that investigated how specific cells of the kidney sense hypoxia25–28, revealing that in response to hypoxia, these cells could increase their production of erythropoietin (EPO) through activation of HIF signalling29–33. This process amplified erythrocyte production to increase the oxygen-carrying capacity of the blood and to counteract a systemic decrease in pO2.
Subsequent studies demonstrated that all cells are armed with HIF and the associated capacity to drive a transcriptional adaptive response that is functionally executed by the expression of proteins that promote a series of responses34–40. These responses include metabolic adaptation, angiogenic switches and vasoactive responses that promote cell and organism survival under hypoxic conditions and stimulate increases in tissue oxygen levels, to overcome the causative hypoxic insult. Although HIF is the most frequently studied hypoxia-sensitive transcription factor and the focus of this Review, multiple other transcriptional pathways display hypoxia sensitivity41,42 and may contribute to the orchestration of the overall transcriptional response to hypoxia42.
The HIF pathway
Canonical HIF regulation
As mentioned above, the HIF pathway is evolutionarily conserved across metazoans1. The oxygen-sensitive nature of HIF has been well described and robustly studied in many cell types3,24,28. Briefly, the transcription factor HIF1α (encoded by HIF1A) is constitutively produced at a high level in most cell types, whereas HIF2α (encoded by EPAS1) is expressed in a more tissue-restricted manner. In the presence of sufficient levels of molecular oxygen (as experienced in normoxia), HIF α-subunits are hydroxylated on defined prolyl residues by three 2-oxoglutarate-dependent prolyl hydroxylase domain-containing proteins (PHD1, PHD2 and PHD3, encoded by EGLN2, EGLN1 and EGLN3, respectively), which leads to HIFα ubiquitination by the von Hippel–Lindau (VHL) protein-recruited E3 ubiquitin ligase complex and proteasomal degradation (Fig. 1). Hydroxylation by the 2-oxoglutarate-dependent factor inhibiting HIF (FIH, encoded by HIF1AN) renders the HIF α-subunits unable to bind to their transcriptional co-activators, the histone acetyl transferases CBP and p300, preventing the formation of a functional transcriptional complex and thereby repressing HIF activity in an oxygen-dependent manner43. Under conditions of hypoxia, however, the enzymatic activity of the PHD and FIH HIF hydroxylases is reduced, thereby stabilizing the HIF α-subunits and increasing their transcriptional activity, respectively. The stabilized HIF α-subunits can translocate to the nucleus where they bind hypoxia response elements (HREs) within regulatory DNA regions of HIF-responsive genes to enhance transcription44. Several hundred genes are known to be HIF-responsive, including EPO, VEGF and CA9. Notably, HIF1 and HIF2 control the expression of discreet yet overlapping gene cohorts3,45. The temporal and quantitative nature of the canonical HIF response in individual cell types is variable and depends on the degree and duration of hypoxic exposure as well as the expression and isoforms of components of the oxygen sensing pathway.
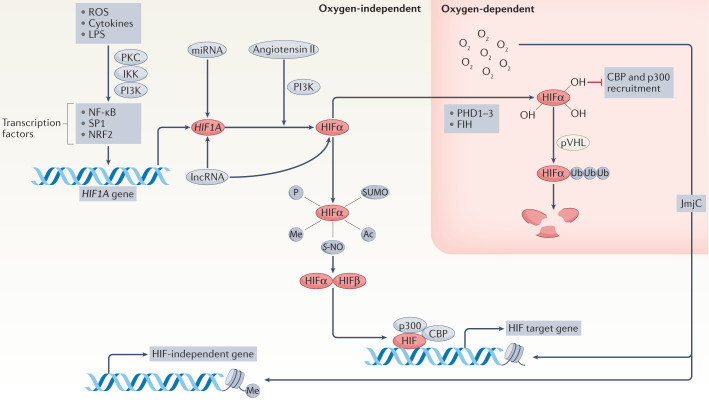
Fig. 1. Mechanisms that regulate HIF.
The activity of the dimeric transcription factor hypoxia-inducible factor (HIF) is regulated in a complex manner through both oxygen-dependent and oxygen-independent mechanisms directed towards the HIF α-subunit. Oxygen-independent mechanisms include the regulation of HIFA gene expression, through the actions of transcription factors such as nuclear factor-κB (NF-κB), specific protein 1 (SP1) and NF-E2-related factor 2 (NRF2), which in turn can be regulated by reactive oxygen species (ROS), cytokine and/or lipopolysaccharide (LPS)-dependent signalling via pathways involving protein kinase C (PKC), inhibitor of NF-κB kinase (IKK) and/or phosphoinositide 3-kinase (PI3K). HIFA mRNA levels and/or translation can be regulated by microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and/or by angiotensin II-mediated signalling involving PI3K. The stability and/or activity of HIFα can be modulated by a variety of oxygen-independent post-translational modifications, including phosphorylation, sumoylation (SUMO), acetylation (Ac), S-nitrosylation (S-NO) and methylation (Me), although the exact contribution of each of these modifications remains to be clarified. The best characterized mechanism of HIFα regulation occurs in an oxygen-dependent manner through the cellular oxygen sensors prolyl hydroxylase domain-containing proteins (PHDs) and the asparagine hydroxylase factor inhibiting HIF (FIH). In normoxia, hydroxylation of two distinct HIFα proline residues by PHD is recognized by the E3 ubiquitin ligase adaptor von Hippel–Lindau (VHL) protein, leading to HIFα poly-ubiquitination and its subsequent degradation. FIH-catalysed hydroxylation of a single HIFα asparagine prevents the recruitment of the adaptor proteins p300 and CBP, which reduces HIF transactivation activity. Under conditions of hypoxia, molecular oxygen is no longer available to the PHDs and FIH, resulting in HIFα stabilization and increased transactivation activity, which leads to the formation of the dimeric HIF transcription factor and the enhanced transcription of HIF target genes. An additional oxygen-dependent regulation occurs via Jumonji C (JmjC) domain-containing histone demethylases, which use molecular oxygen to regulate histone methylation and thereby hypoxia-dependent gene expression. Thus, the activity of the HIF transcription factor is regulated in a complex manner, which allows for a variability of the temporal and dynamic nature of the HIF-dependent response to hypoxia adjusted to the specific needs of each individual cell type.
Additional mechanisms of HIF regulation
Although the ubiquitous, oxygen-dependent canonical HIF pathway has been robustly demonstrated in all investigated cell types, over the past decade, studies have shown that it can also be modified by additional inputs46 (FigS 1 and 2). These findings are important as the existence of such inputs enables qualitatively, quantitatively and temporally distinct patterns of the HIF response in specific cells and contexts. Indeed, strong evidence now demonstrates that a number of inputs beyond oxygen levels are important in shaping the temporally and spatially dynamic nature of the HIF-dependent response to hypoxia, and provide a level of signalling complexity that permits individual cells to set their own sensitivity to oxygen levels. As outlined below, the levels and activity of HIF can be regulated through a number of transcriptional, translational and post-translational mechanisms beyond oxygen-dependent regulation of HIF α-subunit stability.
Fig. 2. Biological signals that regulate HIFα.
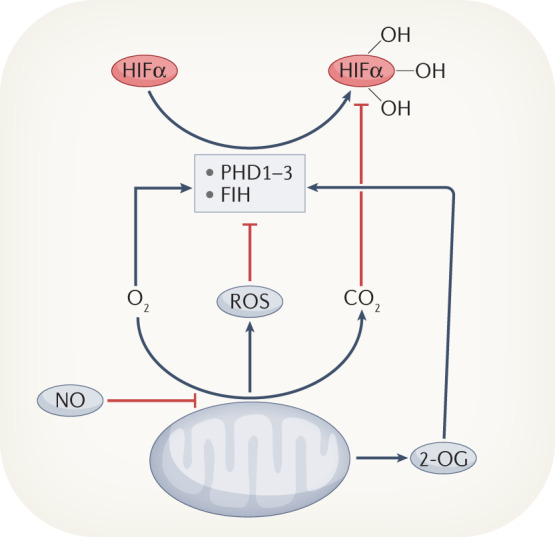
In addition to the oxygen-dependent functions of prolyl hydroxylase domain-containing proteins 1–3 (PHD1–3) and factor inhibiting HIF (FIH), multiple other inputs can modulate hypoxia-inducible factor α (HIFα) stability and/or activity. These include reactive oxygen species (ROS), the physiological gases NO, CO2 and H2S, and the HIF hydroxylase cofactor 2-oxoglutarate (2-OG).
Transcriptional regulation of HIF
At a transcriptional level, the production of HIF1Α mRNA is controlled by a number of transcription factors. Although basal rates of HIF1Α transcription are generally high, differences in transcription rates can be mediated in part by altering the occupation of SP1 binding sites in the HIF1Α promoter by transcription factors that recognize cis-acting elements within the 5′ UTR47.
Stimuli such as ROS can induce transcription of HIF1A mRNA expression in a manner dependent on the activation of the NF-κB pathway48. Angiotensin II (ANGII) increases HIF1A mRNA expression in vascular smooth muscle cells via ROS-dependent activation of the PI3 kinase and PKC pathways49. In support of this finding, ROS increased HIF1Α expression through PI3K and PKC-dependent mechanisms in hypoxic tumour cells under conditions of oxidative stress50. Inflammatory mediators and bacterial products such as lipopolysaccharide can also stimulate HIF1Α and EPAS1 transcription via NF-κB-dependent pathways51–53. The antioxidant transcription factor NRF2 also regulates HIF1A mRNA expression in normoxia and more markedly in the context of reoxygenation following hypoxia54. STAT3 — a transcription factor with wide-ranging roles in processes such as development and inflammation — further regulates HIF1Α expression55. Thus, beyond the well-described oxygen-dependent regulation of HIFα protein stability in cells, transcription of HIF genes is regulated through a number of mechanisms.
Post-transcriptional regulation of HIF
MicroRNAs are short non-coding RNAs that destabilize and/or inhibit the expression and/or translation of their target mRNAs. A subgroup of microRNAs — referred to as HypoxamiRs — regulate the hypoxic response in cells56. For example, the hypoxia-inducible HypoxamiR miR-155 targets HIF1Α57, whereas miR-30c-2-3p and miR-30a-3p target EPAS1 (ref.58). miR-122 is induced by ischaemia and upregulates HIF levels through downregulation of PHD1 (ref.59). The role of miRNAs in sculpting the spatial and temporal nature of the hypoxic response has been reviewed elsewhere60,61.
Long non-coding RNAs (lncRNAs) are another large and diverse family of non-coding RNAs. Like microRNAs, lncRNAs are also involved in the regulation of transcriptional, post-transcriptional and translational events62. Modification of the HIF pathway by lncRNAs has been reported in multiple studies. For example, the HIF1α antisense lncRNA (HIFAL) has an important positive feedforward role in the maintenance of HIF1 signalling63. A myeloid-specific extracellular vesicle-packed HIF1A lncRNA (which works by blocking the interaction between PHD2 and HIF1α, thereby stabilizing HIF1α) has been proposed to promote glycolysis in breast cancer cells64. Similarly, SNHG lncRNA promotes tumour metastasis through HIF stabilization and direct activation of HIF-dependent genes65. HIF1α antisense RNA 2 (HIF1A-AS2) is a hypoxia-inducible lncRNA that is present in mesenchymal glioblastoma multiforme cells and is important in the maintenance of stemness in hypoxic niches66. These few examples implicate important and extensive roles for non-coding RNAs in modulating the HIF response to hypoxia.
Translational regulation of HIF
In addition to the regulation of HIF at the transcriptional level, the translation of HIFα proteins can also be controlled67. For example, ANGII increases ROS–PI3K-mediated translation of HIFα49. Although much remains to be learned about the control of HIFα translation, a number of specific pharmacological inhibitors, including topoisomerase, mTOR and PI3K inhibitors, have been shown to interfere with HIF translation, implicating these pathways in the control of this process67.
Post-translational modifications
Hydroxylation and the resultant ubiquitination of HIF1α via the VHL-recruited E3 ligase complex in the canonical pathway are the primary post-translational modifications responsible for the oxygen-dependent regulation of HIF stability. Several other post-translational modifications of HIF α-subunits have been proposed as potential regulatory mechanisms68. Small ubiquitin-like modifier (SUMO) proteins, which mediate sumoylation, demonstrate sensitivity to hypoxia and can modify HIF69–75. However, the effect of sumoylation on HIF1α stability and activity remains controversial. Other post-translational modifications of HIF1α, including phosphorylation, acetylation, S-nitrosylation and methylation, have been extensively reported and can also influence the stability and activity of HIF α-subunits68. Therefore, the stabilized HIFα protein is heavily decorated with post-translational modifications, which likely differ in nature and degree between cell types to enable fine tuning of the HIF response in specific cell types and contexts. Importantly, post-translational modification by modifiers such as ubiquitin and SUMO are also counter-regulated by the activity of deubiquitinases and sentrin-specific proteases (SENPs), respectively76,77.
Transcriptional complex formation
The HIF pathway is also regulated through the formation of the HIF transcriptional complex — a process that requires assembly of the HIF1α and HIF2α subunits with their transcriptional co-activators (CBP and p300) and recruitment of RNA polymerase 2. As mentioned earlier, the affinity of the HIF α-subunit for CBP and p300 is controlled by the HIF hydroxylase FIH, whereby FIH-mediated hydroxylation of an asparagine residue in HIFα reduces the affinity of HIF for CBP and p300 (refs38–40,43,78,79).
Binding and recruitment of HIF to target genes
HIF-mediated enhancement of target gene transcription requires the presence of at least one HRE, but is also heavily influenced by other factors, including epigenetic modifications, DNA accessibility, chromatin structure and the presence of additional transcription factors45,80–82. The relevance of such additional regulatory factors in the control of HIF target gene selectivity is highlighted by the fact that approximately only 1,000 functional HREs exist despite the HRE DNA sequence occurring more than 1 million times within the human genome80. Studies from the past few years have furthered our understanding of the regulation of gene expression in response to hypoxia with the finding that hypoxia can inhibit 2-oxoglutarate-dependent hydroxylases other than the PHDs, including the Jumonji C (JmjC) domain-containing histone demethylases KDM5A and KDM6A. Inhibition of these demethylases increases histone methylation and thereby typically facilitates hypoxia-dependent gene expression by loosening the chromatin structure80,83,84. The extent to which this process facilitates the transcriptional response to HIF activation remains unclear, but may be of key importance in sculpting the overall cell type-specific transcriptional response to hypoxia.
Regulation of HIF by other biological signals
In addition to the mechanisms outlined above, the overall level of HIF activity in a specific cell type or context can be modulated by a number of other cellular signals, including physiological gases other than oxygen (for example, nitric oxide (NO), carbon dioxide (CO2) and hydrogen sulphide (H2S)), ROS and the availability of HIF hydroxylase co-factors, including Fe2+ and 2-oxoglutarate (also known as α-ketoglutarate) (Fig. 2).
NO is a gasotransmitter that is important for the control of vascular tone, and thus tissue perfusion and overall blood pressure85. However, another important biological role for NO is in the regulation of mitochondrial oxygen consumption by controlling oxygen availability to HIF hydroxylases, and thereby affecting the overall HIF response85,86. Under conditions of hypoxia, whereby PHDs are inhibited and HIF is activated, NO-mediated inhibition of cytochrome c oxidase (COX; also known as complex IV) of the mitochondrial respiratory chain leads to a redistribution of oxygen to the HIF hydroxylases, resulting in increased HIF hydroxylation and subsequent ubiquitination and degradation of HIF85,86.
CO2 is another physiological gas that is generated in mitochondria as the primary gaseous product of respiration. It suppresses HIF1α stability by reducing intracellular pH, which promotes HIF1α degradation via an alternative lysosomal pathway87. Other products of metabolism, including ROS, H2S, iron and tricarboxylic acid (TCA) cycle metabolites (including 2-oxoglutarate, succinate and citrate) modulate the HIF response through the regulation of PHD or FIH activity88–96. Thus, although the availability of oxygen provides the primary signal that determines levels of HIF activity in a cell (via HIF hydroxylase activity), multiple other biological signals can shape the quantitative, spatial and temporal nature of the overall response, thereby providing for a mechanism with which to fine tune the hypoxic response in a cell type- and context-specific manner.
Implications of HIF for metabolism
The HIF pathway has an important role in controlling the expression of genes involved in the regulation of cellular metabolism. We now understand that the metabolic function of a cell, as well as being central to cellular bioenergetics, is also a key driver of cellular behaviour. This function seems to be of particular importance in rapidly proliferative cells, such as leukocytes during an immune response and endothelial cells during angiogenesis. This function is also likely to be of major importance for cells with a high turnover, such as intestinal epithelial cells and for cells that experience physiological hypoxia, such as those in some regions of the kidney. Furthermore, these insights raise the intriguing question as to whether pathological alterations in tumour cell metabolism, which fuels tumour cell proliferation, harness the same physiological processes used by rapidly reproducing immune, endothelial and epithelial cells to alter their metabolism when a period of rapid growth is required. Therefore, better understanding of the mechanisms by which HIF activity regulates cellular metabolism is of key importance to understand both cellular bioenergetics and the control of cell function. Here, we summarize the role of HIF in the metabolic regulation of cells and processes in the human body, which forms the foundation of our emerging understanding of the importance of HIF-dependent metabolic changes in rapidly dividing immune cells.
Metabolic processes affected by HIF activity
HIFs transcriptionally regulate genes involved in energy metabolism under both physiological and pathological conditions, and HIF activity has been associated with metabolic processes such as the Pasteur effect97 and the Warburg effect98. Under normoxic conditions, the majority of cellular ATP is produced by oxidative phosphorylation in the mitochondria — a process that uses approximately 90% of the available oxygen99. HIF-dependent regulation of cellular energy metabolism serves to adjust ATP production when oxygen is limited, thus enabling cell and tissue survival. HIF adapts energy metabolism by enhancing the expression of metabolic enzymes, many of which display an HIF1 (over HIF2) isoform specificity100. HIF1 is therefore considered to be the main driver of metabolic adaptation to hypoxia12. These HIF1-dependent genes control cellular glucose utilization and decrease oxygen consumption to reduce metabolic dependence on oxygen while maintaining appropriate ATP concentrations. In addition, HIF can reduce mitochondrial mass by increasing mitophagy and decreasing mitochondrial biogenesis. Therefore, HIF1 is a major regulator of cellular metabolic strategy during periods in which oxygen availability is limited (Fig. 3). Interestingly, some HIF hydroxylases can also regulate metabolic processes through mechanisms that are at least in part independent of HIF101–104.
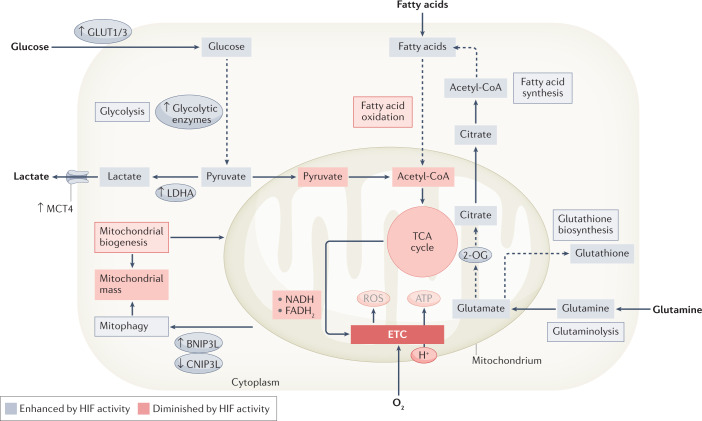
Fig. 3. HIF-mediated adaptations of cellular metabolic pathways.
Hypoxia-inducible factor (HIF) activity has pronounced effects on cellular metabolism, adjusting cellular glucose, fatty acid and glutamine utilization to decrease metabolic oxygen consumption while maintaining appropriate concentrations of ATP and other necessary metabolic intermediates and products. Under hypoxic conditions, increased HIF activity therefore decreases mitochondrial oxygen consumption and mitochondrial mass, and increases glycolysis, fatty acid synthesis and glutaminolysis. Upregulation of glutathione biosynthesis enhances cellular resistance against oxidative stress. 2-OG, 2-oxoglutarate; BNIP3, BCL-2/adenovirus E1B 19-kDa interacting protein 3; ETC, electron transport chain; GLUT, glucose transporter; LDHA, lactate dehydrogenase A; MCT4, monocarboxylate transporter 4; ROS, reactive oxygen species; TCA, tricarboxylic acid cycle.
Glycolysis
Under normoxic conditions, glycolysis provides the first step in the oxidative metabolism of glucose and converts glucose to pyruvate, which is transported into the mitochondria and used for the production of acetyl-CoA. Acetyl-CoA is then fed into the TCA cycle to generate electrons for the ETC in the form of NADH and FADH2. The ETC establishes a proton gradient across the inner mitochondrial membrane, which ATP synthase uses to produce ATP. In hypoxia, HIF1 increases the expression of pyruvate dehydrogenase kinase 1 (PDK1)105,106. This enzyme inhibits the mitochondrial enzyme pyruvate dehydrogenase (PDH) by phosphorylation, preventing the conversion of pyruvate into acetyl-CoA, thereby limiting its availability for the TCA cycle. This represses mitochondrial oxygen consumption, and redirects pyruvate towards glycolysis105,106. This switch from oxidative to glycolytic metabolism effectively diminishes the necessary cellular oxygen availability for ATP production. Furthermore, the reduction of the mitochondrial oxygen consumption leads to reduced mitochondrial production of ROS100, protecting the cell from ROS-induced damage.
Glycolysis is less efficient in terms of ATP production per glucose molecule than oxidative metabolism. To prevent a bioenergetic crisis, increased HIF activity therefore enhances glucose flux through glycolysis by increasing the expression of the glucose transporters GLUT1 (encoded by SLC2A1)107–109 and GLUT3 (encoded by SLC2A3)107,110, which increases cellular glucose uptake. Moreover, HIF1 increases the expression of all glycolytic enzymes100,107 (Table 1), including hexokinases (HK1 and HK2)111, phosphofructokinases (PFKL and PFKP)112,113, aldolases (ALDA and ALDC)112,114, glyceraldehyde 3-phosphate dehydrogenase (GAPDH)115, phosphoglycerate kinase 1 (PGK1)112, enolases (ENO1 and ENO2)114 and pyruvate kinase M (PKM)116. Whether hypoxia also drives a spatial reorganization or compartmentalization of the glycolytic pathway into a so-called metabolon to enhance glycolytic metabolism remains to be determined. Finally, HIF1 enhances the expression of lactate dehydrogenase A (LDHA)114,117 and monocarboxylate transporter 4 (MCT4; encoded by SLC16A3)118. LDHA converts pyruvate to lactate and regenerates NAD+, which is required by GAPDH for additional glycolytic cycles. MCT4 removes lactate from the cell, transporting it into the extracellular space. Therefore, HIF1 drives a switch from oxidative to glycolytic metabolism during periods of hypoxia, an event that has implications not only for cellular bioenergetics but also for cell function and fate.
Table 1.
Glycolytic proteins that are transcriptionally upregulated by hypoxia-inducible factor
| Protein (encoding genes) | Catalysed reaction (per glucose molecule)196 | Refs |
|---|---|---|
| Hexokinases (HK1, HK2) | 1× Glucose phosphorylation utilizing 1× ATP | 107,197 |
| Glucose-6-phosphate isomerase (GPI) | Conversion of 1× glucose-6-phosphate to 1× fructose-6-phosphate | 197–199 |
| Phosphofructokinases (PFKL, PFKP) | Conversion of 1× fructose-6-phosphate to 1× fructose-1,6-bisphosphate utilizing ATP | 107,112,113,197 |
| Aldolases (ALDA, ALDC) | Splitting of 1× fructose-1,6-bisphosphate into 1× dihydroxyacetone phosphate and 1× glyceraldehyde-3-phosphate | 107–109,112,114,197 |
| Triosephosphate isomerase (TPI) | Conversion of 1× dihydroxyacetone phosphate to 1× d-glyceraldehyde-3-phosphate | 97,107 |
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | Conversion of 2× glyceraldehyde 3-phosphate to 2× 1,3-bisphosphoglycerate producing 2× NADH + 2× H+ | 97,107,197 |
| Phosphoglycerate kinase 1 (PGK1) | Conversion of 2× 1,3-bisphosphoglycerate to 2× 3-phosphoglycerate producing 2× ATP | 97,107–109,112,197 |
| Phosphoglycerate mutase (PGAM1) | Conversion of 2× 3-phosphoglycerate to 2× 2-phosphoglycerate | 197 |
| Enolases (ENO1, ENO2) | Conversion of 2× 2-phosphoglycerate to 2× phosphoenolpyruvate producing 2× H2O | 107,112,114,197 |
| Pyruvate kinase M (PKM) | Conversion of 2× phosphoenolpyruvate to 2× pyruvate producing 2× ATP | 107,112,197 |
| Other regulated proteins that support increased glycolytic flux | ||
| Glucose transporters 1 and 3 (GLUT1, GLUT3) | Cellular glucose uptake | 107–110 |
| Lactate dehydrogenase A (LDHA) | Conversion of 2× pyruvate and 2× NADH + 2× H+ to 2× lactate and 2× NAD+ | 107,109,114,117 |
| Monocarboxylate transporter 4 (MCT4) | Cellular lactate efflux | 118 |
Oxidative phosphorylation
While enhancing glycolysis, HIF1 activity also reduces oxidative phosphorylation. HIF1 activity can decrease mitochondrial oxygen consumption by reducing mitochondrial mass through various mechanisms. For instance, enhanced HIF-1 activity promotes mitophagy119,120 by increasing the expression of BCL-2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3) and BNIP3-like (BNIP3L)121,122. HIF1 also reduces mitochondrial biogenesis123 by increasing the expression of MAX interactor 1 (MXI1)123,124. This transcriptional repressor negatively regulates expression of the transcription factor MYC, which decreases MYC-mediated expression of peroxisome proliferator-activated receptor-γ coactivator 1β (PGC1β)123. PGC1β regulates mitochondrial biogenesis and its decrease reduces mitochondrial mass. HIF1 also modifies the mitochondrial ECT — a series of four protein complexes that transfers electrons received from NADH and FADH2 to generate ATP through oxidative phosphorylation. COX is the final electron acceptor of the ECT and consumes the largest amount of oxygen. Under normoxic conditions, COX contains the subunit COX4-1. Under hypoxic conditions, HIF1 activity enhances the expression of the alternative COX subunit COX4-2 and of the mitochondrial protease LON, which degrades the COX4-1 subunit125. Thus, HIF activity changes the COX subunit composition, which may increase the electron transfer efficiency and optimize mitochondrial respiration under hypoxic conditions125. Therefore, hypoxia-induced increases in HIF1 activity reduce oxygen consumption by COX, which in turn likely reduces the production of mitochondrial ROS — a by-product of oxidative phosphorylation.
Fatty acid metabolism
Fatty acid oxidation (FAO) generates acetyl-CoA for the TCA cycle through cyclical shortening of fatty acids by two carbon units per cycle, yielding acetyl-CoA, NADH and FADH2. The last cycle generates two acetyl-CoA molecules. HIF activity indirectly decreases the expression of the medium-chain acyl-CoA dehydrogenase (MCAD) and of the long-chain acyl-CoA dehydrogenase (LCAD)126, which are involved in the first reaction of FAO; thus, decreased levels of MCAD and/or LCAD suppress FAO. A second mechanism of HIF-mediated reduction of FAO has been described in clear-cell renal cell carcinoma, involving inhibition of carnitine palmitoyltransferase 1 A (CPT1A) expression127. CPT1A is a key protein in mitochondrial fatty acid transport; reductions in CPT1A levels lower FAO substrate availability and thus decrease FAO127.
HIF1 activity can also enhance the expression of the VLDL receptor (VLDLR), increasing LDL and HDL uptake128. HIF1 activity further upregulates the expression of insulin-induced gene 2 protein (INSIG2), facilitating the degradation of the enzyme 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase129, which catalyses the rate-limiting step of cholesterol synthesis.
HIFs have also been associated with lipogenesis, including fatty acid synthesis (FAS). FAS requires acetyl-CoA as a substrate, but the above-mentioned HIF1-mediated increase in PDK1 reduces cellular acetyl-CoA levels. To counteract the PDK1-mediated reduction in acetyl-CoA, glutaminolysis — the reductive carboxylation of glutamine — generates 2-oxoglutarate, which is either oxidized to succinate as part of the TCA cycle or used by isocitrate dehydrogenase 1 (IDH1) and IDH2 to produce isocitrate, which is further converted to citrate by aconitase. The ATP citrate lyase (ACLY) converts citrate into acetyl-CoA, which fuels FAS. Increased HIF activity promotes acetyl-CoA production from glutamine via IDHs to fuel lipogenesis130,131. HIF activity can also decrease the activity of the oxoglutarate dehydrogenase complex (OGDH; the TCA cycle enzyme that oxidizes 2-oxoglutarate to succinyl-CoA) likely by enhancing the expression of SIAH2, an E3 ubiquitin ligase that targets a key OGDH subunit for proteasomal degradation132. A decrease in the OGDH activity increases the likelihood that 2-oxoglutarate is converted in the TCA cycle in the reverse direction into isocitrate and citrate, for its utilization for FAS.
Thus, increased HIF activity suppresses FAO, reducing mitochondrial oxygen consumption and thereby adjusting cellular metabolism to meet conditions imposed by limited oxygen availability. In addition, HIF activity enhances lipogenesis, which is necessary to supply fatty acids for membrane synthesis (required for cell growth and proliferation), for the production of signalling molecules and for storage. Hence, HIF plays an important but complex role in the regulation of fatty acid metabolism.
Glutaminolysis and glutathione biosynthesis
An intermediate metabolite of glutaminolysis — a process for the conversion of glutamine to 2-oxoglutarate — is glutamate, and HIF1 activity increases intracellular glutamate levels by enhancing the expression of glutamine transporters133–135 and glutamine-converting glutaminases133,136,137. Glutamate is necessary for the production of glutathione, which protects cells against oxidative stress. HIF1 may drive glutathione generation not only by increasing the availability of glutamate but also by directly enhancing the expression of a glutamate–cysteine ligase modifier subunit (GCLM) — a regulatory subunit of the glutamate-cysteine ligase, which is involved in glutathione biosynthesis138. Thus, HIF1 activity promotes the cellular resilience against oxidative stress.
Amino acid metabolism
HIF1 activity also has an important role in the metabolism of other amino acids139. HIF1 represses levels of the TCA cycle enzyme succinate dehydrogenase subunit A (SDHA) and SDHB140,141, decreasing not only TCA cycle activity but also limiting the production of oxaloacetate. In addition, levels of glutamic-oxaloacetic transaminase 1 (GOT1) and GOT2 are suppressed in an HIF1-dependent manner140. The GOT enzymes convert oxaloacetate into aspartate, thereby reducing aspartate biosynthesis. Furthermore, HIF1 activity affects serine and one-carbon metabolism. One-carbon metabolism supports many physiological processes, including the generation of NADPH; it is mediated by folate metabolism and uses the amino acid serine as a fuel142. HIF activity can increase serine synthesis from glycolytic intermediates by enhancing the expression of the three enzymes in the serine synthesis pathway100. In addition, mitochondrial one-carbon metabolism is upregulated in a HIF-dependent manner by increasing the expression of enzymes such as serine hydroxymethyltransferase 2 (SHMT2) and methylene tetra-hydrofolate dehydrogenase 2 (MTHFD2) that upregulate the production of mitochondrial NADPH, which in turn can fuel glutathione biosynthesis100.
Pentose phosphate pathway
The pentose phosphate pathway (PPP) is connected to the glycolysis pathway, using the glycolytic intermediate glucose-6-phosphate as substrate to generate ribulose-5-phosphate, protons and NADPH via an oxidative and a non-oxidative arm143. Ribulose-5-phosphate can be used to produce nucleic acids or in the non-oxidative PPP arm to generate glyceraldehyde-3-phosphate and fructose-6-phosphate, which are fed back into the glycolysis pathway143. NADPH is required for a variety of processes, including FAS, signalling, and in cellular antioxidant defence via glutathione. Glucose-6-phosphate dehydrogenase (G6PD), which catalyses the first step of the PPP that diverts glucose-6-phosphate away from glycolysis, is upregulated in response to hypoxia in some cancer cells144. However, it remains unclear whether this response is regulated by HIF. By contrast, hypoxia represses G6PD expression in breast cancer cells145. In addition, HIF can regulate the expression of transketolase, an enzyme involved in the non-oxidative arm, which increases glucose flux through this part of the PPP146. Thus, HIF is likely to contribute to the regulation of the PPP, but the detailed mechanism(s) may be cell-type specific and needs further investigation.
These examples and many others make clear that HIF — particularly HIF1 — is a major regulator of cellular metabolic strategy under conditions in which oxygen is limited. Although these effects are driven at the cellular level through alterations in gene expression, they have profound implications for metabolism at the organism level.
Role of HIF in human organismal metabolism
A large part of our understanding of the specific role of HIF in the regulation of metabolic processes has been derived from analyses of cultured cells. Although more difficult to assess, insight into the role of HIF in integrated whole-body energy metabolism in humans can be acquired from the study of patients with VHL mutations, resulting in a higher constitutive level of HIF activity. For example, the autosomal-recessive VHL mutation c.598 C >T reduces the ability of VHL to bind to hydroxylated HIF1α, leading to HIF1α stabilization, and increasing its transcriptional activity147. In patients, this mutation causes Chuvash polycythaemia, which is characterized by increased haemoglobin and haematocrit levels, and abnormalities in cardiopulmonary function. Analyses of metabolic markers in these patients reveal increased blood lactate levels and acidosis in the skeletal muscle during exercise148. The expression of GLUT1 and glycolytic enzymes was also upregulated147,148, supporting the notion that glycolysis is enhanced in patients with increased HIF1 activity.
A 2020 report described a patient with a different VHL mutation (c.222 C >A, p.V74V)149. This mutation differed from the mutation underlying Chuvash polycythaemia and led to a reduced amount of VHL, thereby increasing HIF1α levels149. The patient presented with familial erythrocytosis type 2, but also with metabolic alterations that partly differed from those typical of Chuvash polycythaemia149. Persistent hypoglycaemia was observed alongside changes in carbohydrate and lipid metabolism, combined with decreased skeletal muscle mitochondrial respiration, decreased FAO and uncoupling of ATP production from oxygen consumption149. The expression of several HIF target genes was enhanced, including those encoding GLUT1, BNIP3L and MXI1. Analysis of the metabolome demonstrated increased glycolytic flux and decreased TCA cycle activity. Findings also indicated increased glutamine and altered aspartate metabolism. Seven additionally analysed amino acids were decreased in the blood, likely because they were used as a carbon source. HDL levels were also reduced, indicating alterations in lipid metabolism. The patient further exhibited a higher-than-normal ratio of oxidized-to-reduced glutathione levels149, which may be indicative of increased oxidative stress. Alternatively, glutathione levels may be upregulated by HIF to enhance the cellular defence against future oxidative stress. As described earlier, increased HIF activity generally occurs during hypoxia, which if resolved, is followed by re-oxygenation. In the context of ischaemia–reperfusion injury, oxidative stress caused by re-oxygenation is a major contributor to tissue injury. Thus, glutathione levels may be increased to attenuate the damage otherwise inflicted by oxidative stress in the likely event of re-oxygenation.
The role of HIF in the regulation of energy metabolism also has clinical relevance in the context of HIF PHD inhibitors (PHIs), which have now been approved for the treatment of renal anaemia. Clinical studies of roxadustat and daprodustat reported decreased triglycerides, total serum cholesterin, LDL and HDL cholesterol following PHI treatment150.
Therefore, VHL mutations or pharmacological hydroxylase inhibition in humans manifest as changes in human metabolism that broadly reflect the role of HIF in metabolism at the cellular level. The variability in the metabolic effects between patients with different VHL mutations or with HIF hydroxylase inhibitor treatment may be explained by the extent to which HIFα is stabilized. VHL also regulates proteins other than HIFα151 and some of the metabolic effects observed in patients with VHL mutations may thus not be solely dependent on HIFα stabilization. Of note, PHI administration to patients with renal anaemia leads to lower EPO peak levels than achieved with erythropoiesis-stimulating agents and EPO is considered to be one of the most sensitive genes for HIF activity150. The amount of PHI administered may therefore only induce a subset of HIF-regulated genes in some patients and in selected organs. This area, especially the potential additional effects of PHIs, will need further analyses in the future.
Implications of HIF-mediated changes in cell metabolism
As outlined above, HIF activation is tightly controlled in a cell-type and context-specific way with a complex array of inputs determining overall HIF activity in an individual cell at any given time. This mechanism enables temporal and spatial heterogeneity of the HIF response across a tissue. As a major regulator of cellular metabolism, HIF has a key role in maintaining cellular bioenergetic homeostasis, which enables highly energy-dependent processes, such as cellular growth and proliferation, to continue under hypoxic conditions. These metabolic effects of HIF also have important consequences in pathological situations. It is well recognized that tumour cells require an alternative to oxidative phosphorylation to support their growth, which is achieved primarily through increased glycolysis, even in the presence of available oxygen (a process known as the Warburg effect or aerobic glycolysis). The pathological role of HIF in tumour cell metabolism is profound100,152 and is not further discussed here. Instead, we focus on a physiological role for HIF and its metabolic effects in rapidly dividing cells of the immune system during immune and inflammatory responses. The diverse roles of HIF in the control of immune cell function have been covered elsewhere5,153–156, and so here we focus on more recent findings — mainly from the past few years — on the role of HIF in regulating immune cell phenotype and function, and the physiological consequences of these effects for infection and inflammation.
Implications for immune cell function
When activated, immune cells typically undergo rapid increases in activity and expansion to induce an effective immune response157. This process is highly energy dependent, and requires cells to have the capacity to alter their metabolic strategy to maintain sufficient levels of ATP to support this process. Furthermore, active inflammation is a highly metabolically demanding process and as a result, sites of inflammation are often profoundly hypoxic158. Therefore, resident and infiltrating immune cells are exposed to steep oxygen gradients and can experience hypoxia while at the same time needing to proliferate and function. As described earlier, oxidative metabolism is compromised under hypoxic conditions, leading to cellular reliance on oxygen-independent pathways, such as glycolysis, to provide ATP157. We now appreciate that HIF has an important role not only in the control of immune cell phenotype and function5,153,154,159 but also in the regulation of immune cell metabolism, thereby acting as a key regulator of immunometabolism154 (Fig. 4).
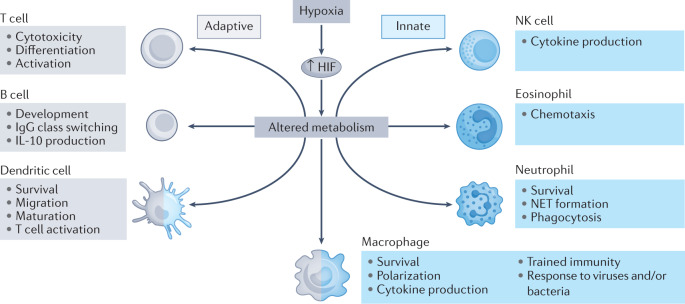
Fig. 4. HIF-dependent regulation of immunometabolism.
Hypoxia increases hypoxia-inducible factor (HIF) activity, which affects the phenotype and function of innate and adaptive immune cells, including epithelial cells, neutrophils, macrophages, dendritic cells, T and B lymphocytes, natural killer (NK) cells and innate lymphoid cells through regulation of immune cell metabolism. NET, neutrophil extracellular trap.
Since the first report of a role for HIF in the control of macrophage function in 2003 (ref.160), multiple studies have demonstrated that HIF contributes to the regulation of all cell types involved in the innate and adaptive immune response, including epithelial cells, neutrophils, macrophages, dendritic cells, T cells, B cells, natural killer cells and innate lymphoid cells155,161. A consistent theme of these studies is that a clear relationship exists between HIF and immunometabolism. Of particular note, the degree to which a cell depends on glycolysis for ATP production seems to be associated with the phenotype in a cell type-specific manner.
For example, activation of HIF1 has been shown to promote glycolysis and phenotypic changes in innate immune cells, such as neutrophils, macrophages and dendritic cells. In neutrophils, the switch to glycolysis is associated with the promotion of survival, phagocytic capacity and neutrophil extracellular trap formation154, thereby enhancing their innate immune activity. In macrophages, the promotion of glycolysis is associated with polarization to an M1 pro-inflammatory phenotype as opposed to an M2 anti-inflammatory phenotype, which uses oxidative phosphorylation for ATP generation162,163. Other macrophage functions affected by HIF activation include survival, cytokine production, trained immunity and their response to viruses and bacteria154. HIF-dependent changes in dendritic cells that have been associated with altered cellular metabolism include effects on cell survival, migration, maturation and T cell activation164. HIF activation also promotes eosinophil migration, although whether this effect is a consequence of alterations in metabolism remains unclear165.
HIF activation also increases glycolysis in adaptive immune cells. HIF-mediated glycolysis is associated with the differentiation and development of T cells and B cells. In T cells, the HIF-dependent induction of glycolysis has been associated with the activation and differentiation of different T cell subtypes, as well as effects on T cell cytotoxicity, and memory154. In B cells, HIF-dependent changes in glycolysis are associated with B cell development, IgG class switching and IL-10 production154. In natural killer cells, HIF has been associated with altered cytokine production and improved wound healing166.
More recent studies have expanded our understanding of the effects of HIF on immune cell function, with the identification of roles for HIF in innate lymphoid cell plasticity in the gut167, regulatory T cell abundance168 and neutrophil motility169. These studies further support a central role for HIF in immunity that is heavily cell-type and context specific. As outlined below, our understanding of how these cell-type specific roles of HIF relate to the relevance of HIF in overall inflammation and immunity (which of course involves many immune cell types) is evolving.
A growing body of research implicates a role for HIF in the overall physiological immune response to infection and pathological inflammation.
Effects of HIF on infection
A number of studies have investigated the impact of inflammation and infection in mice with conditional deletion of HIF pathway components in immune cells. Mice with myeloid cell-specific knockdown of HIF1Α display increased susceptibility to infection with Mycobacterium tuberculosis156,170. HIF was also found to be protective in a zebrafish model of tuberculosis171. Similarly, the presence of HIF1 in macrophages contributed to the induction of pro-inflammatory genes in response to Helicobacter pylori infection, whereas myeloid cell-specific deletion of HIF1A in mice led to worsening of inflammation and gastritis following Helicobacter pylori colonization172. HIF also has a key role in host defence against uropathogenic Escherichia coli infection173. Thus, HIF activity in immune cells seems to be largely associated with a protective response to pathogenic infections.
Effects of HIF on inflammation
The anti-inflammatory effects of HIF activation have been extensively demonstrated5,11,154. This understanding suggests that pharmacological activation of HIF may represent a potentially important new approach to the control of inflammation5,11,154. Indeed, pharmacological activation of HIF using PHIs has demonstrated anti-inflammatory effects in a range of in vivo models5. This finding is supported by results from phase 1B clinical trial for the treatment of ulcerative colitis, in which protective effects of treatment with the PHI GB004 were reported174. A 2018 study demonstrated that deficiency of HIF in B cells increased susceptibility to collagen-induced arthritis, suggesting a key role of HIF for B cell function in the context of autoimmunity175. However, in some inflammatory conditions such as colitis, HIF isoforms may exert opposing roles. For example, HIF1α expressed in intestinal epithelial cells is protective in colitis176 whereas HIF2α is detrimental177,178. Thus, the role of HIF in inflammation is complex, although evidence to date suggests that activation of the HIF pathway is largely anti-inflammatory.
Implications for PHD therapeutics
As we have described, HIF is a central regulator of human cellular and organism metabolism. Interestingly, living under hypoxia at high altitude has been associated with a decreased prevalence of obesity and type 2 diabetes in humans179–181. This observation raises the question of whether increasing HIF activity with use of PHIs could be harnessed as a novel treatment option for metabolic disorders. A possible beneficial effect of PHI treatment in metabolic disorders is supported by the observation that use of PHIs in patients with renal anaemia can decrease levels of triglycerides, total serum cholesterin, and LDL and HDL cholesterol150. Similarly, in mice, PHI treatment decreased levels of serum cholesterin and circulating fatty acids182,183, and attenuated the deterioration of glucose tolerance induced by a high-fat diet183. PHI treatment also protected obese mice against the development of atherosclerosis184. Thus, available evidence supports the notion that pharmacological activation of HIF can induce metabolic reprogramming and suggests that PHIs may represent a novel treatment option for metabolic disorders and their adverse sequelae.
The activation of HIF also has profound implications for the function of immune cells through the regulation of immunometabolic processes5,153,154. Preclinical studies, predominantly in models of intestinal inflammation, have largely demonstrated PHIs to be strongly anti-inflammatory153 and as a consequence, pharmacological PHIs are now being explored for the treatment of colitis in patients174,185. Interestingly, a key mechanism by which PHIs seem to provide protection in colitis is through the promotion of intestinal epithelial barrier function. This barrier protective mechanism likely works in combination with effects on immune cell function186,187. Thus, pharmacological efficacy of PHIs in diseases such as colitis might be enhanced by using drug delivery techniques to target the tissue of interest188. Furthermore, combining PHIs with more traditional anti-inflammatory therapeutics may have added benefits in inflammatory disease.
The kidney experiences an oxygen gradient, which is likely a prerequisite for its function as an oxygen-sensing organ9,189; however, the kidney is also frequently exposed to pathophysiological hypoxia as a consequence of, for example, inflammatory processes, ischaemia and/or fibrosis in the context of acute kidney injury, CKD and in specific conditions such as systemic lupus erythematosus20,190,191. Furthermore, injury or disease-mediated changes in metabolic processes in proximal tubule epithelial cells (which are influenced by HIF) may also affect the susceptibility of these cells to damage in response to stressors192–195. On the other hand, the fact that preclinical analyses have mostly demonstrated a protective effect of pharmacological HIF hydroxylase inhibition in murine models of CKD, must be taken into account20. By contrast, genetic modifications of proteins of the HIF pathway in murine glomerular cells, renal epithelial cells or endothelial cells have more often been detrimental than protective in murine CKD models20. Thus, genetic modulations of the HIF pathway in specific cell types may not necessarily reflect the outcome of systemic PHI administration. These points need to be kept in mind when considering the long-term nephrological implications of PHI treatment for patients with renal anaemia.
Conclusions
The role of HIF in the cellular, tissue and organismal adaptation to hypoxia remains an important and highly active field. Of particular and growing interest are the metabolic consequences of HIF and their effects on immune cell phenotype and function. The multiple inputs into the HIF pathway provide individual cell types with the ability for nuanced and context-specific HIF responses to microenvironmental situations of hypoxia. Furthermore, individual leukocyte sub-types respond to hypoxia with cell-type specific changes in their cellular metabolism with consequences for immune cell function. Our understanding of the full effects of HIF-dependent changes on immune cell metabolism and associated changes in cell function is in its infancy, but some themes are emerging, such as the consequences associated with a change to glycolytic metabolism on macrophage and lymphocyte phenotype. However, little is currently known about how hypoxia-dependent changes in other metabolic pathways can influence immune cell function and the (patho)physiological consequences of this for organs such as the kidney. This is an exciting and important emerging area of research that will have major implications for our understanding of immune responses.
Several areas relevant to our understanding of the importance of HIF-dependent changes in metabolism in immune cell function require further research. These include the impact of HIF activation in in vivo models of inflammatory disease for which HIF activators such as PHIs have been found to be strongly anti-inflammatory. Indeed, phase 2 clinical trials studying the potential use of hydroxylase inhibitors in inflammatory bowel disease are ongoing174,185.
With respect to our understanding of the regulators of the HIF pathway in immune cells, an area in its infancy is our understanding of the role of non-coding RNAs in their regulation of the HIF pathway. Indeed, the complex nature of the multiple inputs into HIF signalling implicate the potential use of mathematical and systems biology approaches to understanding the spatial, temporal and quantitative nature of the HIF response in individual cells and contexts. Finally, an area of controversy and importance with respect to our understanding of the role of oxygen sensing in general is whether PHIs might also affect non-HIF targets. The near future should provide the answers to these fascinating and important questions.
Glossary
- Cis-acting elements
Regions of non-coding DNA that regulate the transcription of genes.
- Sumoylation
Post-translational protein modification by attachment of SUMO (small ubiquitin-related modifier) proteins, involving the formation of isopeptide bonds with ε-amino groups of acceptor Lys residues of target proteins.
- Pasteur effect
The increased glucose utilization for anaerobic ATP production in response to diminished oxygen availability.
- Warburg effect
The preferential use of aerobic glycolysis rather than oxidative phosphorylation by tumour cells for energy production.
- Mitophagy
The selective degradation of mitochondria by autophagy.
- One-carbon metabolism
A series of interlinking metabolic pathways that provide 1 C units (methyl groups) for the synthesis of nucleotides, polyamines, amino acids, creatine and phospholipids.
Author contributions
The authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Nephrology thanks Holger Eltzschig, Masaomi Nangaku, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
C.T.T. has consulted for Gossamer Bio and Akebia. C.C.S. declares no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Taylor CT, McElwain JC. Ancient atmospheres and the evolution of oxygen sensing via the hypoxia-inducible factor in metazoans. Physiology. 2010;25:272–279. doi: 10.1152/physiol.00029.2010. [DOI] [PubMed] [Google Scholar]
- 2.Wenger RH, Kurtcuoglu V, Scholz CC, Marti HH, Hoogewijs D. Frequently asked questions in hypoxia research. Hypoxia. 2015;3:35–43. doi: 10.2147/HP.S92198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Taylor CT. Mitochondria and cellular oxygen sensing in the HIF pathway. Biochem. J. 2008;409:19–26. doi: 10.1042/BJ20071249. [DOI] [PubMed] [Google Scholar]
- 5.Taylor CT, Colgan SP. Regulation of immunity and inflammation by hypoxia in immunological niches. Nat. Rev. Immunol. 2017;17:774–785. doi: 10.1038/nri.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Provot S, Schipani E. Fetal growth plate: a developmental model of cellular adaptation to hypoxia. Ann. NY Acad. Sci. 2007;1117:26–39. doi: 10.1196/annals.1402.076. [DOI] [PubMed] [Google Scholar]
- 7.Schödel J, Ratcliffe PJ. Mechanisms of hypoxia signalling: new implications for nephrology. Nat. Rev. Nephrol. 2019;15:641–659. doi: 10.1038/s41581-019-0182-z. [DOI] [PubMed] [Google Scholar]
- 8.Scholz H, et al. Kidney physiology and susceptibility to acute kidney injury: implications for renoprotection. Nat. Rev. Nephrol. 2021;17:335–349. doi: 10.1038/s41581-021-00394-7. [DOI] [PubMed] [Google Scholar]
- 9.Nolan KA, Wenger RH. Source and microenvironmental regulation of erythropoietin in the kidney. Curr. Opin. Nephrol. Hypertens. 2018;27:277–282. doi: 10.1097/MNH.0000000000000420. [DOI] [PubMed] [Google Scholar]
- 10.Lindholm ME, Rundqvist H. Skeletal muscle hypoxia-inducible factor-1 and exercise. Exp. Physiol. 2016;101:28–32. doi: 10.1113/EP085318. [DOI] [PubMed] [Google Scholar]
- 11.Pham K, Parikh K, Heinrich EC. Hypoxia and inflammation: insights from high-altitude physiology. Front. Physiol. 2021;12:676782. doi: 10.3389/fphys.2021.676782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schito L, Semenza GL. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annu. Rev. Physiol. 2014;76:39–56. doi: 10.1146/annurev-physiol-021113-170322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 16.Ryan S, Taylor CT, McNicholas WT. Selective activation of inflammatory pathways by intermittent hypoxia in obstructive sleep apnea syndrome. Circulation. 2005;112:2660–2667. doi: 10.1161/CIRCULATIONAHA.105.556746. [DOI] [PubMed] [Google Scholar]
- 17.Prabhakar NR, Peng YJ, Nanduri J. Hypoxia-inducible factors and obstructive sleep apnea. J. Clin. Invest. 2020;130:5042–5051. doi: 10.1172/JCI137560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fine LG, Orphanides C, Norman JT. Progressive renal disease: the chronic hypoxia hypothesis. Kidney Int. Suppl. 1998;65:S74–S78. [PubMed] [Google Scholar]
- 19.Honda T, Hirakawa Y, Nangaku M. The role of oxidative stress and hypoxia in renal disease. Kidney Res. Clin. Pract. 2019;38:414–426. doi: 10.23876/j.krcp.19.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faivre A, Scholz CC, de Seigneux S. Hypoxia in chronic kidney disease: towards a paradigm shift? Nephrol. Dial. Transpl. 2021;36:1782–1790. doi: 10.1093/ndt/gfaa091. [DOI] [PubMed] [Google Scholar]
- 21.Shirvaliloo M. The blood-gas barrier in COVID-19: an overview of the effects of SARS-CoV-2 infection on the alveolar epithelial and endothelial cells of the lung. Tissue Barriers. 2021;9:937013. doi: 10.1080/21688370.2021.1937013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simonson TS, et al. Silent hypoxaemia in COVID-19 patients. J. Physiol. 2021;599:1057–1065. doi: 10.1113/JP280769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortega-Saenz P, Lopez-Barneo J. Physiology of the carotid body: from molecules to disease. Annu. Rev. Physiol. 2020;82:127–149. doi: 10.1146/annurev-physiol-020518-114427. [DOI] [PubMed] [Google Scholar]
- 24.Cummins EP, Strowitzki MJ, Taylor CT. Mechanisms and consequences of oxygen and carbon dioxide sensing in mammals. Physiol. Rev. 2020;100:463–488. doi: 10.1152/physrev.00003.2019. [DOI] [PubMed] [Google Scholar]
- 25.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 28.Fandrey J, Schödel J, Eckardt KU, Katschinski DM, Wenger RH. Now a Nobel gas: oxygen. Pflugers Arch. 2019;471:1343–1358. doi: 10.1007/s00424-019-02334-8. [DOI] [PubMed] [Google Scholar]
- 29.Imeri F, et al. Generation of renal Epo-producing cell lines by conditional gene tagging reveals rapid HIF-2 driven Epo kinetics, cell autonomous feedback regulation, and a telocyte phenotype. Kidney Int. 2019;95:375–387. doi: 10.1016/j.kint.2018.08.043. [DOI] [PubMed] [Google Scholar]
- 30.Koury MJ, Haase VH. Anaemia in kidney disease: harnessing hypoxia responses for therapy. Nat. Rev. Nephrol. 2015;11:394–410. doi: 10.1038/nrneph.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi H, et al. Distinct subpopulations of FOXD1 stroma-derived cells regulate renal erythropoietin. J. Clin. Invest. 2016;126:1926–1938. doi: 10.1172/JCI83551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broeker KAE, et al. Prolyl-4-hydroxylases 2 and 3 control erythropoietin production in renin-expressing cells of mouse kidneys. J. Physiol. 2022;600:671–694. doi: 10.1113/JP282615. [DOI] [PubMed] [Google Scholar]
- 33.Dahl SL, et al. Fate-mapping of erythropoietin-producing cells in mouse models of hypoxaemia and renal tissue remodelling reveals repeated recruitment and persistent functionality. Acta Physiol. 2022;234:e13768. doi: 10.1111/apha.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell PH, Pugh CW, Ratcliffe PJ. Inducible operation of the erythropoietin 3’ enhancer in multiple cell lines: evidence for a widespread oxygen-sensing mechanism. Proc. Natl Acad. Sci. USA. 1993;90:2423–2427. doi: 10.1073/pnas.90.6.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc. Natl Acad. Sci. USA. 1993;90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaakkola P, et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 37.Ivan M, et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 38.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hewitson KS, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 40.Lando D, et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cummins EP, Comerford KM, Scholz C, Bruning U, Taylor CT. Hypoxic regulation of NF-κB signaling. Methods Enzymol. 2007;435:479–492. doi: 10.1016/S0076-6879(07)35025-8. [DOI] [PubMed] [Google Scholar]
- 42.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch. 2005;450:363–371. doi: 10.1007/s00424-005-1413-7. [DOI] [PubMed] [Google Scholar]
- 43.Volkova YL, Pickel C, Jucht AE, Wenger RH, Scholz CC. The asparagine hydroxylase FIH — a unique oxygen sensor. Antioxid. Redox Signal. 2022 doi: 10.1089/ars.2022.0003. [DOI] [PubMed] [Google Scholar]
- 44.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci. STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 45.Smythies JA, et al. Inherent DNA-binding specificities of the HIF-1α and HIF-2α transcription factors in chromatin. EMBO Rep. 2019;20:e46401. doi: 10.15252/embr.201846401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ivan M, Kaelin WG., Jr The EGLN-HIF O2-sensing system: multiple inputs and feedbacks. Mol. Cell. 2017;66:772–779. doi: 10.1016/j.molcel.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minet E, et al. HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5’UTR. Biochem. Biophys. Res. Commun. 1999;261:534–540. doi: 10.1006/bbrc.1999.0995. [DOI] [PubMed] [Google Scholar]
- 48.Bonello S, et al. Reactive oxygen species activate the HIF-1α promoter via a functional NFκB site. Arterioscler. Thromb. Vasc. Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 49.Pagé EL, Robitaille GA, Pouysségur J, Richard DE. Induction of hypoxia-inducible factor-1alpha by transcriptional and translational mechanisms. J. Biol. Chem. 2002;277:48403–48409. doi: 10.1074/jbc.M209114200. [DOI] [PubMed] [Google Scholar]
- 50.Koshikawa N, Hayashi J, Nakagawara A, Takenaga K. Reactive oxygen species-generating mitochondrial DNA mutation up-regulates hypoxia-inducible factor-1α gene transcription via phosphatidylinositol 3-kinase-Akt/protein kinase C/histone deacetylase pathway. J. Biol. Chem. 2009;284:33185–33194. doi: 10.1074/jbc.M109.054221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Uden P, Kenneth NS, Rocha S. Regulation of hypoxia-inducible factor-1α by NF-κB. Biochem. J. 2008;412:477–484. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Uden P, et al. Evolutionary conserved regulation of HIF-1β by NF-κB. PLoS Genet. 2011;7:e1001285. doi: 10.1371/journal.pgen.1001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frede S, Stockmann C, Freitag P, Fandrey J. Bacterial lipopolysaccharide induces HIF-1 activation in human monocytes via p44/42 MAPK and NF-κB. Biochem. J. 2006;396:517–527. doi: 10.1042/BJ20051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potteti HR, et al. Nrf2 mediates hypoxia-inducible HIF1α activation in kidney tubular epithelial cells. Am. J. Physiol. Renal Physiol. 2021;320:F464–F474. doi: 10.1152/ajprenal.00501.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niu G, et al. Signal transducer and activator of transcription 3 is required for hypoxia-inducible factor-1α RNA expression in both tumor cells and tumor-associated myeloid cells. Mol. Cancer Res. 2008;6:1099–1105. doi: 10.1158/1541-7786.MCR-07-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertero T, Rezzonico R, Pottier N, Mari B. Impact of microRNAs in the cellular response to hypoxia. Int. Rev. Cell Mol. Biol. 2017;333:91–158. doi: 10.1016/bs.ircmb.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 57.Bruning U, et al. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1α activity during prolonged hypoxia. Mol. Cell Biol. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mathew LK, et al. Restricted expression of miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas enhances HIF2α activity. Cancer Discov. 2014;4:53–60. doi: 10.1158/2159-8290.CD-13-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ju C, et al. Hypoxia-inducible factor-1α-dependent induction of miR122 enhances hepatic ischemia tolerance. J. Clin. Invest. 2021;131:e140300. doi: 10.1172/JCI140300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gee HE, Ivan C, Calin GA, Ivan M. HypoxamiRs and cancer: from biology to targeted therapy. Antioxid. Redox Signal. 2014;21:1220–1238. doi: 10.1089/ars.2013.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol. Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 63.Zheng F, et al. The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis. Nat. Commun. 2021;12:1341. doi: 10.1038/s41467-021-21535-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen F, et al. Extracellular vesicle-packaged HIF-1α-stabilizing lncRNA from tumour-associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat. Cell Biol. 2019;21:498–510. doi: 10.1038/s41556-019-0299-0. [DOI] [PubMed] [Google Scholar]
- 65.Xu L, et al. LncRNA SNHG11 facilitates tumor metastasis by interacting with and stabilizing HIF-1α. Oncogene. 2020;39:7005–7018. doi: 10.1038/s41388-020-01512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mineo M, et al. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masoud GN, Li W. HIF-1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm. Sin. B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Albanese A, Daly LA, Mennerich D, Kietzmann T, Sée V. The role of hypoxia-inducible factor post-translational modifications in regulating its localisation, stability, and activity. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms22010268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Comerford KM, et al. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl Acad. Sci. USA. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shao R, et al. Increase of SUMO-1 expression in response to hypoxia: direct interaction with HIF-1α in adult mouse brain and heart in vivo. FEBS Lett. 2004;569:293–300. doi: 10.1016/j.febslet.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 71.Bae SH, et al. Sumoylation increases HIF-1α stability and its transcriptional activity. Biochem. Biophys. Res. Commun. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- 72.Berta MA, Mazure N, Hattab M, Pouysségur J, Brahimi-Horn MC. SUMOylation of hypoxia-inducible factor-1α reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 2007;360:646–652. doi: 10.1016/j.bbrc.2007.06.103. [DOI] [PubMed] [Google Scholar]
- 73.Carbia-Nagashima A, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1α during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 74.Nunez-O’Mara A, Berra E. Deciphering the emerging role of SUMO conjugation in the hypoxia-signaling cascade. Biol. Chem. 2013;394:459–469. doi: 10.1515/hsz-2012-0319. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, et al. HIF1alpha protein SUMOylation is an important protective mechanism of action of hypothermia in hypoxic cardiomyocytes. Mol. Med. Rep. 2021;23:476. doi: 10.3892/mmr.2021.12115. [DOI] [PubMed] [Google Scholar]
- 76.Mennerich D, Kubaichuk K, Kietzmann T. DUBs, hypoxia, and cancer. Trends Cancer. 2019;5:632–653. doi: 10.1016/j.trecan.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Günter J, Ruiz-Serrano A, Pickel C, Wenger RH, Scholz CC. The functional interplay between the HIF pathway and the ubiquitin system — more than a one-way road. Exp. Cell Res. 2017;356:152–159. doi: 10.1016/j.yexcr.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 78.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 79.McNeill LA, et al. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the beta-carbon of asparagine-803. Biochem. J. 2002;367:571–575. doi: 10.1042/bj20021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kindrick JD, Mole DR. Hypoxic regulation of gene transcription and chromatin: cause and effect. Int. J. Mol. Sci. 2020;21:8320. doi: 10.3390/ijms21218320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Orlando IMC, et al. Distal and proximal hypoxia response elements cooperate to regulate organ-specific erythropoietin gene expression. Haematologica. 2020;105:2774–2784. doi: 10.3324/haematol.2019.236406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schörg A, et al. Destruction of a distal hypoxia response element abolishes trans-activation of the PAG1 gene mediated by HIF-independent chromatin looping. Nucleic Acids Res. 2015;43:5810–5823. doi: 10.1093/nar/gkv506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Batie M, et al. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science. 2019;363:1222–1226. doi: 10.1126/science.aau5870. [DOI] [PubMed] [Google Scholar]
- 84.Chakraborty AA, et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science. 2019;363:1217–1222. doi: 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taylor CT, Moncada S. Nitric oxide, cytochrome C oxidase, and the cellular response to hypoxia. Arterioscler. Thromb. Vasc. Biol. 2010;30:643–647. doi: 10.1161/ATVBAHA.108.181628. [DOI] [PubMed] [Google Scholar]
- 86.Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1α. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 87.Selfridge AC, et al. Hypercapnia suppresses the HIF-dependent adaptive response to hypoxia. J. Biol. Chem. 2016;291:11800–11808. doi: 10.1074/jbc.M116.713941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp. Physiol. 2006;91:807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 89.Semenza GL, Prabhakar NR. The role of hypoxia-inducible factors in carotid body (patho) physiology. J. Physiol. 2018;596:2977–2983. doi: 10.1113/JP275696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flannigan KL, et al. Proresolution effects of hydrogen sulfide during colitis are mediated through hypoxia-inducible factor-1alpha. FASEB J. 2015;29:1591–1602. doi: 10.1096/fj.14-266015. [DOI] [PubMed] [Google Scholar]
- 91.Miles AL, Burr SP, Grice GL, Nathan JA. The vacuolar-ATPase complex and assembly factors, TMEM199 and CCDC115, control HIF1alpha prolyl hydroxylation by regulating cellular iron levels. eLife. 2017;6:e22693. doi: 10.7554/eLife.22693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bailey PSJ, Nathan JA. Metabolic regulation of hypoxia-inducible transcription factors: the role of small molecule metabolites and iron. Biomedicines. 2018;6:60. doi: 10.3390/biomedicines6020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bailey PSJ, et al. ABHD11 maintains 2-oxoglutarate metabolism by preserving functional lipoylation of the 2-oxoglutarate dehydrogenase complex. Nat. Commun. 2020;11:4046. doi: 10.1038/s41467-020-17862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nytko KJ, Spielmann P, Camenisch G, Wenger RH, Stiehl DP. Regulated function of the prolyl-4-hydroxylase domain (PHD) oxygen sensor proteins. Antioxid. Redox Signal. 2007;9:1329–1338. doi: 10.1089/ars.2007.1683. [DOI] [PubMed] [Google Scholar]
- 95.Hewitson KS, et al. Structural and mechanistic studies on the inhibition of the hypoxia-inducible transcription factor hydroxylases by tricarboxylic acid cycle intermediates. J. Biol. Chem. 2007;282:3293–3301. doi: 10.1074/jbc.M608337200. [DOI] [PubMed] [Google Scholar]
- 96.Koivunen P, et al. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 2007;282:4524–4532. doi: 10.1074/jbc.M610415200. [DOI] [PubMed] [Google Scholar]
- 97.Seagroves TN, et al. Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol. Cell Biol. 2001;21:3436–3444. doi: 10.1128/MCB.21.10.3436-3444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Semenza GL. HIF-1 mediates the Warburg effect in clear cell renal carcinoma. J. Bioenerg. Biomembr. 2007;39:231–234. doi: 10.1007/s10863-007-9081-2. [DOI] [PubMed] [Google Scholar]
- 99.Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- 100.Samanta D, Semenza GL. Metabolic adaptation of cancer and immune cells mediated by hypoxia-inducible factors. Biochim. Biophys. Acta Rev. Cancer. 2018;1870:15–22. doi: 10.1016/j.bbcan.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 101.Zhang N, et al. The asparaginyl hydroxylase factor inhibiting HIF-1α is an essential regulator of metabolism. Cell Metab. 2010;11:364–378. doi: 10.1016/j.cmet.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scholz CC, et al. FIH regulates cellular metabolism through hydroxylation of the deubiquitinase OTUB1. PLoS Biol. 2016;14:e1002347. doi: 10.1371/journal.pbio.1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.D’Hulst G, et al. PHD1 controls muscle mTORC1 in a hydroxylation-independent manner by stabilizing leucyl tRNA synthetase. Nat. Commun. 2020;11:174. doi: 10.1038/s41467-019-13889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yoon H, et al. PHD3 loss promotes exercise capacity and fat oxidation in skeletal muscle. Cell Metab. 2020;32:215–228.e7. doi: 10.1016/j.cmet.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 106.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 107.Iyer NV, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maltepe E, Schmidt JV, Baunoch D, Bradfield CA, Simon MC. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature. 1997;386:403–407. doi: 10.1038/386403a0. [DOI] [PubMed] [Google Scholar]
- 109.Ryan HE, Lo J, Johnson RS. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.O’Rourke JF, Pugh CW, Bartlett SM, Ratcliffe PJ. Identification of hypoxically inducible mRNAs in HeLa cells using differential-display PCR. Role of hypoxia-inducible factor-1. Eur. J. Biochem. 1996;241:403–410. doi: 10.1111/j.1432-1033.1996.00403.x. [DOI] [PubMed] [Google Scholar]
- 111.Mathupala SP, Rempel A, Pedersen PL. Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J. Biol. Chem. 2001;276:43407–43412. doi: 10.1074/jbc.M108181200. [DOI] [PubMed] [Google Scholar]
- 112.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 1994;269:23757–23763. doi: 10.1016/S0021-9258(17)31580-6. [DOI] [PubMed] [Google Scholar]
- 113.Minchenko O, Opentanova I, Caro J. Hypoxic regulation of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene family (PFKFB-1-4) expression in vivo. FEBS Lett. 2003;554:264–270. doi: 10.1016/S0014-5793(03)01179-7. [DOI] [PubMed] [Google Scholar]
- 114.Semenza GL, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 115.Graven KK, Yu Q, Pan D, Roncarati JS, Farber HW. Identification of an oxygen responsive enhancer element in the glyceraldehyde-3-phosphate dehydrogenase gene. Biochim. Biophys. Acta. 1999;1447:208–218. doi: 10.1016/S0167-4781(99)00118-9. [DOI] [PubMed] [Google Scholar]
- 116.Luo W, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Firth JD, Ebert BL, Ratcliffe PJ. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. J. Biol. Chem. 1995;270:21021–21027. doi: 10.1074/jbc.270.36.21021. [DOI] [PubMed] [Google Scholar]
- 118.Ullah MS, Davies AJ, Halestrap AP. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1α-dependent mechanism. J. Biol. Chem. 2006;281:9030–9037. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- 119.Zhang H, et al. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Bellot G, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo K, et al. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 2001;8:367–376. doi: 10.1038/sj.cdd.4400810. [DOI] [PubMed] [Google Scholar]
- 122.Sowter HM, Ratcliffe PJ, Watson P, Greenberg AH, Harris AL. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 2001;61:6669–6673. [PubMed] [Google Scholar]
- 123.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 124.Corn PG, et al. Mxi1 is induced by hypoxia in a HIF-1-dependent manner and protects cells from c-Myc-induced apoptosis. Cancer Biol. Ther. 2005;4:1285–1294. doi: 10.4161/cbt.4.11.2299. [DOI] [PubMed] [Google Scholar]
- 125.Fukuda R, et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 126.Huang, et al. HIF-1-mediated suppression of acyl-CoA dehydrogenases and fatty acid oxidation is critical for cancer progression. Cell Rep. 2014;8:1930–1942. doi: 10.1016/j.celrep.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 127.Du W, et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat. Commun. 2017;8:1769. doi: 10.1038/s41467-017-01965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shen GM, et al. Hypoxia-inducible factor-1 (HIF-1) promotes LDL and VLDL uptake through inducing VLDLR under hypoxia. Biochem. J. 2012;441:675–683. doi: 10.1042/BJ20111377. [DOI] [PubMed] [Google Scholar]
- 129.Hwang S, et al. Hypoxia-inducible factor 1α activates insulin-induced gene 2 (Insig-2) transcription for degradation of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase in the liver. J. Biol. Chem. 2017;292:9382–9393. doi: 10.1074/jbc.M117.788562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc. Natl Acad. Sci. USA. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sun RC, Denko NC. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 2014;19:285–292. doi: 10.1016/j.cmet.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Corbet C, et al. The SIRT1/HIF2α axis drives reductive glutamine metabolism under chronic acidosis and alters tumor response to therapy. Cancer Res. 2014;74:5507–5519. doi: 10.1158/0008-5472.CAN-14-0705. [DOI] [PubMed] [Google Scholar]
- 134.Morotti M, et al. Hypoxia-induced switch in SNAT2/SLC38A2 regulation generates endocrine resistance in breast cancer. Proc. Natl Acad. Sci. USA. 2019;116:12452–12461. doi: 10.1073/pnas.1818521116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yoo HC, et al. A variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 2020;31:267–283.e12. doi: 10.1016/j.cmet.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 136.Stegen S, et al. HIF-1α promotes glutamine-mediated redox homeostasis and glycogen-dependent bioenergetics to support postimplantation bone cell survival. Cell Metab. 2016;23:265–279. doi: 10.1016/j.cmet.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xiang L, et al. Glutaminase 1 expression in colorectal cancer cells is induced by hypoxia and required for tumor growth, invasion, and metastatic colonization. Cell Death Dis. 2019;10:40. doi: 10.1038/s41419-018-1291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lu H, et al. Chemotherapy triggers HIF-1-dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc. Natl Acad. Sci. USA. 2015;112:E4600–E4609. doi: 10.1073/pnas.1421018112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bouthelier A, Aragones J. Role of the HIF oxygen sensing pathway in cell defense and proliferation through the control of amino acid metabolism. Biochim. Biophys. Acta Mol. Cell Res. 2020;1867:118733. doi: 10.1016/j.bbamcr.2020.118733. [DOI] [PubMed] [Google Scholar]
- 140.Melendez-Rodriguez F, et al. HIF1α suppresses tumor cell proliferation through inhibition of aspartate biosynthesis. Cell Rep. 2019;26:2257–2265.e4. doi: 10.1016/j.celrep.2019.01.106. [DOI] [PubMed] [Google Scholar]
- 141.Dahia PL, et al. A HIF1α regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1:72–80. doi: 10.1371/journal.pgen.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Payen VL, Porporato PE, Baselet B, Sonveaux P. Metabolic changes associated with tumor metastasis, part 1: tumor pH, glycolysis and the pentose phosphate pathway. Cell Mol. Life Sci. 2016;73:1333–1348. doi: 10.1007/s00018-015-2098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gao L, Mejías R, Echevarría M, López-Barneo J. Induction of the glucose-6-phosphate dehydrogenase gene expression by chronic hypoxia in PC12 cells. FEBS Lett. 2004;569:256–260. doi: 10.1016/j.febslet.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 145.Samanta D, et al. PHGDH expression is required for mitochondrial redox homeostasis, breast cancer stem cell maintenance, and lung metastasis. Cancer Res. 2016;76:4430–4442. doi: 10.1158/0008-5472.CAN-16-0530. [DOI] [PubMed] [Google Scholar]
- 146.Zhao F, et al. Imatinib resistance associated with BCR-ABL upregulation is dependent on HIF-1α-induced metabolic reprograming. Oncogene. 2010;29:2962–2972. doi: 10.1038/onc.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ang SO, et al. Disruption of oxygen homeostasis underlies congenital Chuvash polycythemia. Nat. Genet. 2002;32:614–621. doi: 10.1038/ng1019. [DOI] [PubMed] [Google Scholar]
- 148.Formenti F, et al. Regulation of human metabolism by hypoxia-inducible factor. Proc. Natl Acad. Sci. USA. 2010;107:12722–12727. doi: 10.1073/pnas.1002339107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Perrotta S, et al. Effects of germline VHL deficiency on growth, metabolism, and mitochondria. N. Engl. J. Med. 2020;382:835–844. doi: 10.1056/NEJMoa1907362. [DOI] [PubMed] [Google Scholar]
- 150.Haase VH. Hypoxia-inducible factor-prolyl hydroxylase inhibitors in the treatment of anemia of chronic kidney disease. Kidney Int. Suppl. 2021;11:8–25. doi: 10.1016/j.kisu.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang J, Zhang Q. VHL and hypoxia signaling: beyond HIF in cancer. Biomedicines. 2018;6:35. doi: 10.3390/biomedicines6010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee P, Chandel NS, Simon MC. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 2020;21:268–283. doi: 10.1038/s41580-020-0227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Colgan SP, Furuta GT, Taylor CT. Hypoxia and innate immunity: keeping up with the HIFsters. Annu. Rev. Immunol. 2020;38:341–363. doi: 10.1146/annurev-immunol-100819-121537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McGettrick AF, O’Neill LAJ. The role of HIF in immunity and inflammation. Cell Metab. 2020;32:524–536. doi: 10.1016/j.cmet.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 155.Krzywinska E, Stockmann C. Hypoxia, metabolism and immune cell function. Biomedicines. 2018;6:56. doi: 10.3390/biomedicines6020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hammond FR, Lewis A, Elks PM. If it’s not one thing, HIF’s another: immunoregulation by hypoxia inducible factors in disease. FEBS J. 2020;287:3907–3916. doi: 10.1111/febs.15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gaber T, Chen Y, Krauss PL, Buttgereit F. Metabolism of T lymphocytes in health and disease. Int. Rev. Cell Mol. Biol. 2019;342:95–148. doi: 10.1016/bs.ircmb.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 158.Campbell EL, et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity. 2014;40:66–77. doi: 10.1016/j.immuni.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Taylor CT, Doherty G, Fallon PG, Cummins EP. Hypoxia-dependent regulation of inflammatory pathways in immune cells. J. Clin. Invest. 2016;126:3716–3724. doi: 10.1172/JCI84433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cramer T, et al. HIF-1α is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/S0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sadiku P, Walmsley SR. Hypoxia and the regulation of myeloid cell metabolic imprinting: consequences for the inflammatory response. EMBO Rep. 2019;20:e47388. doi: 10.15252/embr.201847388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wang T, et al. HIF1α-induced glycolysis metabolism is essential to the activation of inflammatory macrophages. Mediat. Inflamm. 2017;2017:9029327. doi: 10.1155/2017/9029327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jantsch J, et al. Hypoxia and hypoxia-inducible factor-1α modulate lipopolysaccharide-induced dendritic cell activation and function. J. Immunol. 2008;180:4697–4705. doi: 10.4049/jimmunol.180.7.4697. [DOI] [PubMed] [Google Scholar]
- 165.Crotty Alexander LE, et al. Myeloid cell HIF-1α regulates asthma airway resistance and eosinophil function. J. Mol. Med. 2013;91:637–644. doi: 10.1007/s00109-012-0986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sobecki M, et al. NK cells in hypoxic skin mediate a trade-off between wound healing and antibacterial defence. Nat. Commun. 2021;12:4700. doi: 10.1038/s41467-021-25065-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Krzywinska E, et al. The transcription factor HIF-1α mediates plasticity of NKp46+ innate lymphoid cells in the gut. J. Exp. Med. 2022;219:e20210909. doi: 10.1084/jem.20210909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Clambey ET, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc. Natl Acad. Sci. USA. 2012;109:E2784–E2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sormendi S, et al. HIF2α is a direct regulator of neutrophil motility. Blood. 2021;137:3416–3427. doi: 10.1182/blood.2020007505. [DOI] [PubMed] [Google Scholar]
- 170.Resende M, et al. Myeloid HIF-1α regulates pulmonary inflammation during experimental Mycobacterium tuberculosis infection. Immunology. 2020;159:121–129. doi: 10.1111/imm.13131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ogryzko NV, et al. Hif-1α-induced expression of Il-1β protects against mycobacterial infection in zebrafish. J. Immunol. 2019;202:494–502. doi: 10.4049/jimmunol.1801139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Matak P, et al. Myeloid HIF-1 is protective in Helicobacter pylori-mediated gastritis. J. Immunol. 2015;194:3259–3266. doi: 10.4049/jimmunol.1401260. [DOI] [PubMed] [Google Scholar]
- 173.Lin AE, et al. Role of Hypoxia inducible factor-1α (HIF-1α) in innate defense against uropathogenic Escherichia coli infection. PLoS Pathog. 2015;11:e1004818. doi: 10.1371/journal.ppat.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Danese S, et al. Randomised clinical trial: a phase 1b study of GB004, an oral HIF-1α stabiliser, for treatment of ulcerative colitis. Aliment. Pharmacol. Ther. 2022;55:401–411. doi: 10.1111/apt.16753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Meng X, et al. Hypoxia-inducible factor-1α is a critical transcription factor for IL-10-producing B cells in autoimmune disease. Nat. Commun. 2018;9:251. doi: 10.1038/s41467-017-02683-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Karhausen J, et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J. Clin. Invest. 2004;114:1098–1106. doi: 10.1172/JCI200421086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Singhal R, Shah YM. Oxygen battle in the gut: hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J. Biol. Chem. 2020;295:10493–10505. doi: 10.1074/jbc.REV120.011188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kerber EL, et al. The importance of hypoxia-inducible factors (HIF-1 and HIF-2) for the pathophysiology of inflammatory bowel disease. Int. J. Mol. Sci. 2020;21:8551. doi: 10.3390/ijms21228551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Voss JD, Masuoka P, Webber BJ, Scher AI, Atkinson RL. Association of elevation, urbanization and ambient temperature with obesity prevalence in the United States. Int. J. Obes. 2013;37:1407–1412. doi: 10.1038/ijo.2013.5. [DOI] [PubMed] [Google Scholar]
- 180.Woolcott OO, et al. Inverse association between altitude and obesity: a prevalence study among Andean and low-altitude adult individuals of Peru. Obesity. 2016;24:929–937. doi: 10.1002/oby.21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Woolcott OO, et al. Inverse association between diabetes and altitude: a cross-sectional study in the adult population of the United States. Obesity. 2014;22:2080–2090. doi: 10.1002/oby.20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Michailidou Z, et al. Adipocyte pseudohypoxia suppresses lipolysis and facilitates benign adipose tissue expansion. Diabetes. 2015;64:733–745. doi: 10.2337/db14-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rahtu-Korpela L, et al. HIF prolyl 4-hydroxylase-2 inhibition improves glucose and lipid metabolism and protects against obesity and metabolic dysfunction. Diabetes. 2014;63:3324–3333. doi: 10.2337/db14-0472. [DOI] [PubMed] [Google Scholar]
- 184.Rahtu-Korpela L, et al. Hypoxia-inducible factor prolyl 4-hydroxylase-2 inhibition protects against development of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2016;36:608–617. doi: 10.1161/ATVBAHA.115.307136. [DOI] [PubMed] [Google Scholar]
- 185.US National Library of Medicine. ClinicalTrials.govhttps://clinicaltrials.gov/ct2/show/NCT04556383 (2022).
- 186.Brown E, Taylor CT. Hypoxia-sensitive pathways in intestinal inflammation. J. Physiol. 2017;596:2985–2989. doi: 10.1113/JP274350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tambuwala MM, et al. Loss of prolyl hydroxylase-1 protects against colitis through reduced epithelial cell apoptosis and increased barrier function. Gastroenterology. 2010;139:2093–2101. doi: 10.1053/j.gastro.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 188.Tambuwala MM, et al. Targeted delivery of the hydroxylase inhibitor DMOG provides enhanced efficacy with reduced systemic exposure in a murine model of colitis. J. Control. Release. 2015;217:221–227. doi: 10.1016/j.jconrel.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 189.Wenger RH, Hoogewijs D. Regulated oxygen sensing by protein hydroxylation in renal erythropoietin-producing cells. Am. J. Physiol. Renal Physiol. 2010;298:F1287–F1296. doi: 10.1152/ajprenal.00736.2009. [DOI] [PubMed] [Google Scholar]
- 190.Nangaku M, Eckardt KU. Hypoxia and the HIF system in kidney disease. J. Mol. Med. 2007;85:1325–1330. doi: 10.1007/s00109-007-0278-y. [DOI] [PubMed] [Google Scholar]
- 191.Chen PM, et al. Kidney tissue hypoxia dictates T cell-mediated injury in murine lupus nephritis. Sci. Transl Med. 2020;12:eaay1620. doi: 10.1126/scitranslmed.aay1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kang HM, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat. Med. 2015;21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dhillon P, et al. The nuclear receptor ESRRA protects from kidney disease by coupling metabolism and differentiation. Cell Metab. 2021;33:379–394.e8. doi: 10.1016/j.cmet.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Hasegawa S, et al. The oral hypoxia-inducible factor prolyl hydroxylase inhibitor enarodustat counteracts alterations in renal energy metabolism in the early stages of diabetic kidney disease. Kidney Int. 2020;97:934–950. doi: 10.1016/j.kint.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 195.Legouis D, et al. Altered proximal tubular cell glucose metabolism during acute kidney injury is associated with mortality. Nat. Metab. 2020;2:732–743. doi: 10.1038/s42255-020-0238-1. [DOI] [PubMed] [Google Scholar]
- 196.Kierans SJ, Taylor CT. Regulation of glycolysis by the hypoxia-inducible factor (HIF): implications for cellular physiology. J. Physiol. 2021;599:23–37. doi: 10.1113/JP280572. [DOI] [PubMed] [Google Scholar]
- 197.Mole DR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yoon DY, et al. Identification of genes differentially induced by hypoxia in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2001;288:882–886. doi: 10.1006/bbrc.2001.5867. [DOI] [PubMed] [Google Scholar]
- 199.Funasaka T, Yanagawa T, Hogan V, Raz A. Regulation of phosphoglucose isomerase/autocrine motility factor expression by hypoxia. FASEB J. 2005;19:1422–1430. doi: 10.1096/fj.05-3699com. [DOI] [PubMed] [Google Scholar]