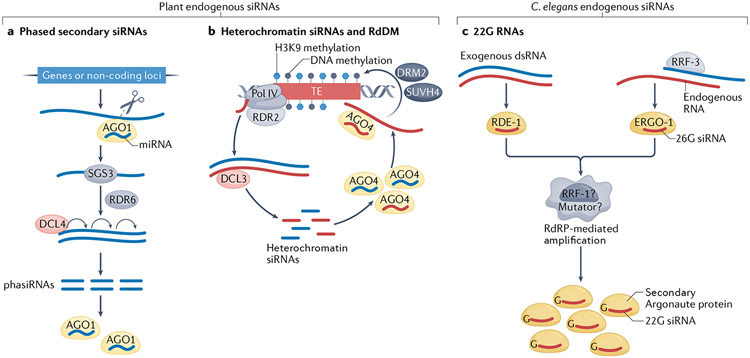
Fig. 2 ∣. Endogenous siRNAs in plants and Caenorhabditis elegans.
a ∣ Phased secondary small interfering RNAs (phasiRNAs) are produced in plants following transcript targeting and cleavage by a microRNA (miRNA)–ARGONAUTE 1 (AGO1) complex, which leads to the production of double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) using the transcript as a template, a process that also requires the RNA-binding protein SUPPRESSOR OF GENE SILENCING 3 (SGS3). The dsRNA is processively cleaved into phasiRNAs by DICER-LIKE 4 (DCL4), starting from the parental RNA end produced by the miRNA-guided cleavage, b ∣ A simplified model of RNA-directed DNA methylation (RdDM). Transposable elements (TEs) in plants produce 24-nucleotide small interfering RNAs (siRNAs) through a process mediated by RNA polymerase IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DCL3; the siRNAs are loaded into AGO4 and target this siRNA-induced silencing complex (siRISC) to nascent transcripts at homologous DNA loci. The siRISC recruits the de novo DNA methyltransferase DRM2 to perform cytosine methylation, and histone methyltransferases such as SUVH4 that methylate histone H3 Lys9 (H3K9), thereby silencing the expression of the TEs. c ∣ In Caenorhabditis elegans, ‘primary’ siRNAs derived from single-stranded RNA or from dsRNA are loaded onto the ‘primary’ Argonaute proteins ERGO-1 or RDE-1, respectively. RRF-3 is an RNA-dependent RNA polymerase (RdRP) that mediates the production of 26-nucleotide-long 5′-guanosine siRNAs (26G siRNAs). The loaded primary Argonaute protein directs the production — possibly in ‘mutator’ foci, where the RdRP RRF-1 is localized — of 22G ‘secondary’ siRNAs, which are loaded onto ‘secondary’ Argonaute proteins to direct gene silencing.