Figure 3. Glycan-based phylogenetic trees.
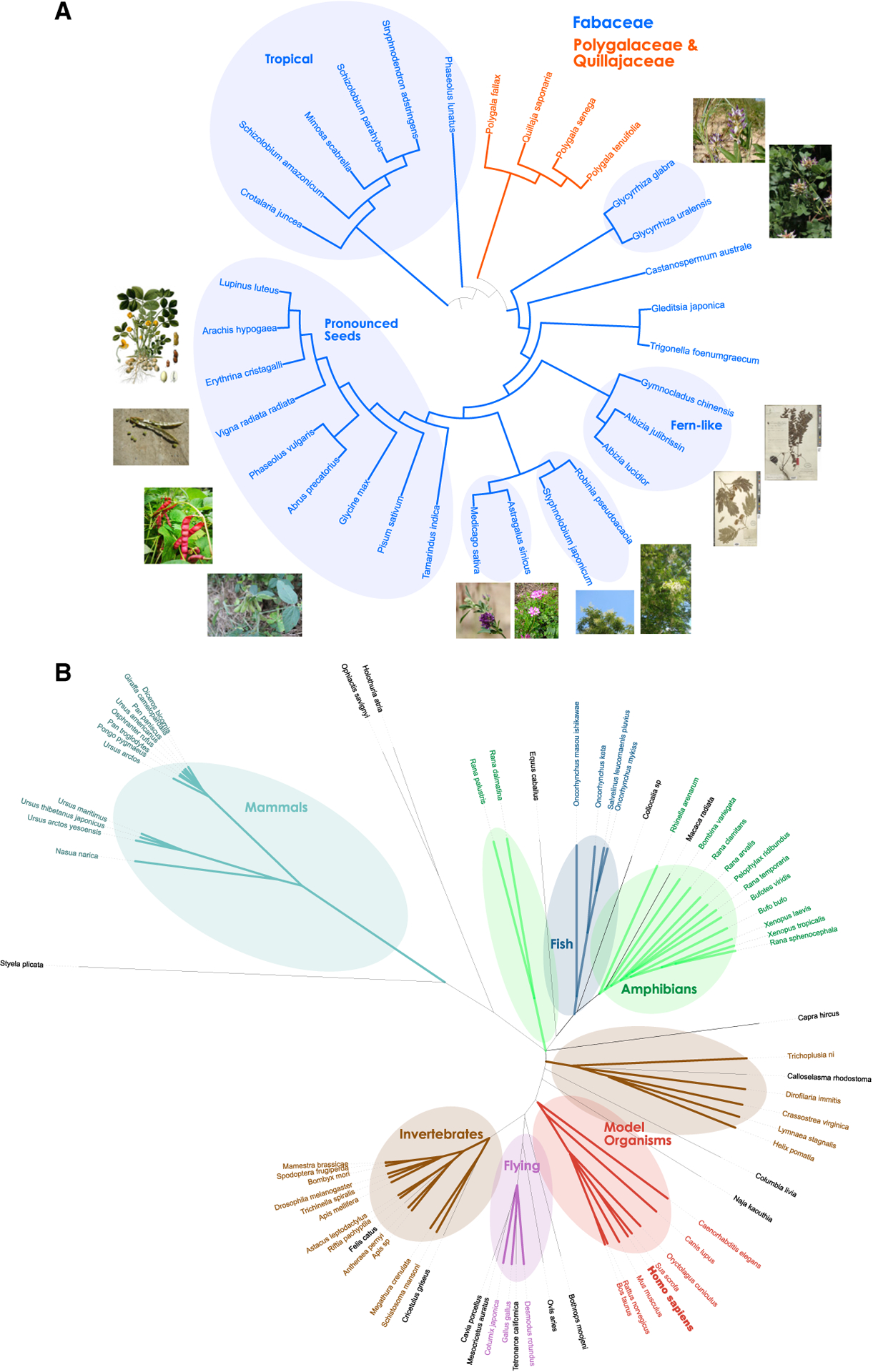
For all 30 species from the order Fabales with at least five glycans (A) or 93 species from the kingdom Animalia with at least two glycans in our dataset (B), we averaged their glycan representations from the species-level SweetNet model, constructed a cosine distance matrix among all species, and performed hierarchical clustering to obtain a dendrogram. The shown phylogenetic tree was drawn with the Interactive Tree of Life v5.5 software (Letunic and Bork, 2019). For (A), we colored species belonging to the taxonomic families Fabaceae and Polygalaceae/Quillajaceae. (A and B) We further annotated clusters enriched for certain groups of plants (A) or animals (B) that shared characteristics. The species Homo sapiens is depicted in bold. Images used for (A) stemmed from the public domain. Exceptions were from Creative Commons licenses requiring attribution and included Astragalus sinicus (https://commons.wikimedia.org/wiki/File:Chinese_milkvetch_Ziyunying.JPG), Glycyrrhiza uralensis (https://commons.wikimedia.org/wiki/File:Glycyrrhiza_uralensis_IMG_1086.jpg), and Glycyrrhiza glabra (https://commons.wikimedia.org/wiki/File:Glycyrrhiza_glabra_Y13.jpg).
