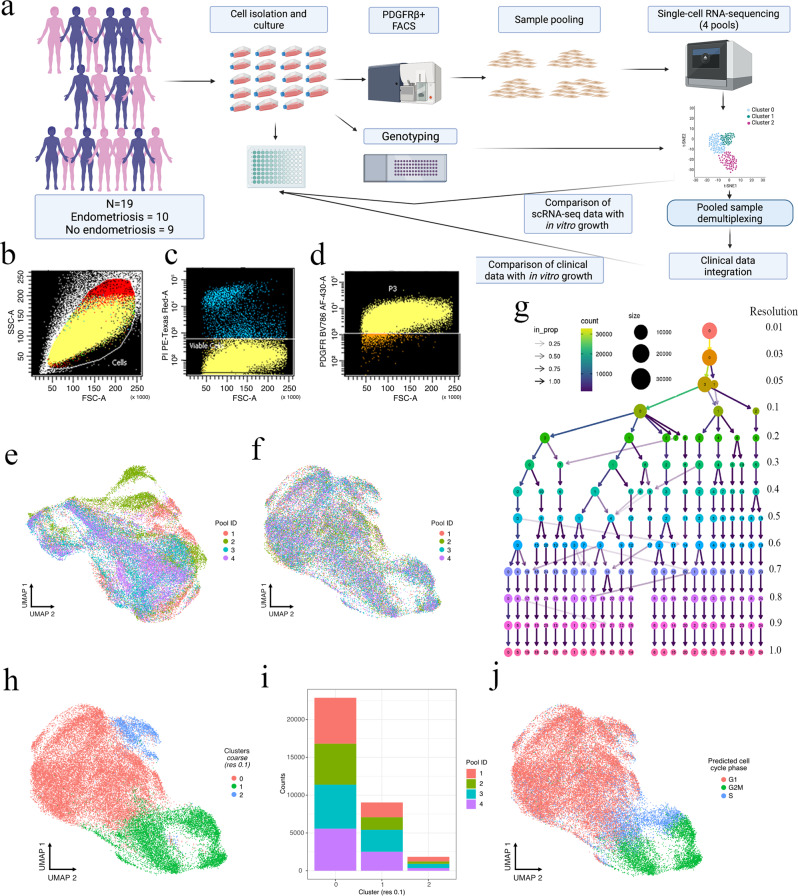
Fig. 1. Experimental design and quality control for high throughput single-cell RNA sequencing of purified endometrial mesenchymal cells.
a Endometrial biopsies were isolated, from 19 different women and cultured. In vitro growth assays, DNA isolation for genotyping and PDGFRβ + FACS purification was performed. Purified cells from nineteen samples were pooled into four lanes and run on the 10X Chromium controller and scRNA-seq data was analysed and cell clustering was performed. Samples were assigned to their source individual and clusters compared to clinical data. Both scRNA-seq data and clinical data were compared to in vitro growth. Mesenchymal stromal cells were purified via FACS (b) forward and side scatter, c viability and d PDGFRβ + expression. UMAP plot distribution of scRNA-seq data was determined both e pre- and f post harmony correction of between-pool variations introduced through technical variations. g Clustree analysis was used to determine the most stable level for cellular clustering. h UMAP plot of integrated scRNA-seq data at a clustering resolution of 0.1 identified three distinct clusters with minimal subsequent mixing at a finer resolution. i At a cluster resolution of 0.1, cell numbers from each pool in each cluster remained stable suggesting clusters were formed from biological differences rather than technical effects. j UMAP plot of scRNA-seq data labelled by cell cycle phase inferred by the CellCycleScoring function in Seurat. a Created with BioRender.com.