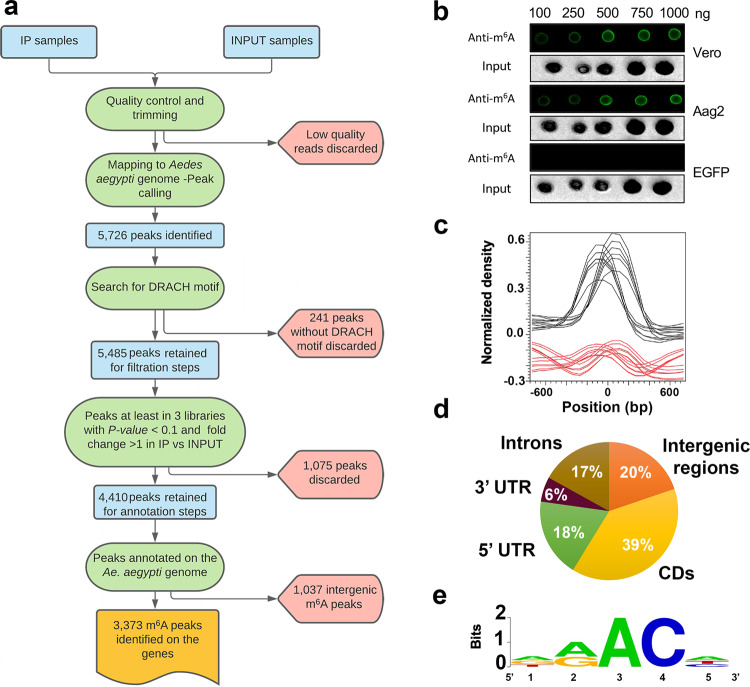
Fig. 1. m6A modification of Ae. aegypti transcripts analysed by dot blot and methylated RNA immunoprecipitation and sequencing (MeRIP-Seq).
a A diagram showing the MeRIP-Seq data analysis procedure. b Confirmation of RNA N6-methyladenosine (m6A) methylation in Ae. aegypti. Total RNA from Aag2 and Vero cells were extracted and subjected to a dot blot assay using a specific anti-m6A antibody. EGFP transcripts were synthetised in vitro and used as negative control. The input RNAs were directly stained with ethidium bromide. c Normalized density of m6A peaks between immunoprecipitated (black lines) and input (red lines) samples following MeRIP-Seq indicating enrichment of m6A in the IP samples as part of the quality control of the data. d Pie chart showing distribution of m6A peaks in Ae. aegypti transcripts regions. e The consensus m6A motif DRA*CH (D = G/A/U, R = G/A, * modified A, H = U/A/C) was enriched in the identified m6A peaks.