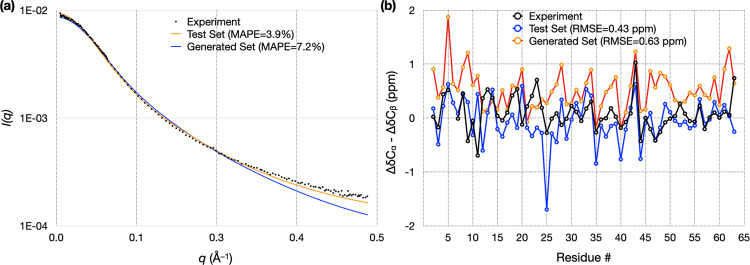
Fig. 7. Validation of autoencoder-generated conformations for ChiZ by experimental SAXS and chemical shift data.
a Comparison of experimental and calculated SAXS profiles. MAPE was calculated as , where and are experimental and predicted scattering intensities, respectively, and denotes the average over all the data points. b Comparison of experimental and calculated secondary chemical shifts. The experimental data and the MD simulations are reported previously14. RMSE was calculated as the square root of . Calculations were done on either the test set comprising 12,180 conformations sampled from 12 MD runs, or on an autoencoder-generated set comprising the same number of conformations, after refinement. The autoencoder was trained on a combined training set comprising 52,200 conformations sampled from the 12 MD runs.