Abstract
Introduction
Advances in the scientific understanding of the skin and characteristic genomic dermal signatures continue to develop rapidly. Nonetheless, skin diagnosis remains predicated on a subjective visual examination, frequently followed by biopsy and histology. These procedures often are not sufficiently sensitive, and in the case of many inflammatory diseases, biopsies are not justified, creating a situation where high-quality samples can be difficult to obtain. The wealth of molecular information available and the pace at which new data are acquired suggest that methods for minimally invasive biomarker collection could dramatically alter our understanding of skin disease and positively impact treatment paradigms.
Methods
A chemical method was optimized to covalently modify custom dermal patches with single-stranded DNA that could bind to messenger RNA. These patches were applied to ex vivo skin samples and penetration evaluated by histological methods. Patches were then applied to both the skin of normal human subjects (lower arm) as well as lesional skin of psoriasis patients, and the transcriptome captured (N = 7; 33 unique samples). Standard RNA-Seq processing was performed to assess the gene detection rate and assessments made of the reproducibility of the extraction procedure as well as the overlap with matched punch biopsy samples from the same patient.
Results
We have developed a dermal biomarker patch (DBP) designed to be minimally invasive and extract the dermal transcriptome. Using this platform, we have demonstrated successful molecular analysis from healthy human skin and psoriatic lesions, replicating the molecular information captured with punch biopsy.
Conclusion
This DBP enables an unprecedented ability to monitor the molecular “fingerprint” of the skin over time or with various interventions, and generate previously inaccessible rich datasets. Furthermore, use of the DBP could be favored by patients relative to biopsy by limiting pain resulting from biopsy procedures. Given the large dynamic range observed in psoriatic skin, analysis of complex phenotypes is now possible, and the power of machine-learning methods can be brought to bear on dermatologic disease.
Keywords: Dermal biomarker patch, Minimally invasive, Next-generation sequencing, Psoriasis, RNA-Seq, Transcriptome
Key Summary Points
| Biomarkers have become a preferred method for disease detection and patient monitoring. Furthermore, the development of robust biomarker sets has brought the promise of precision medicine to the treatment of a number of diseases. |
| Multiple biomarker sets related to skin diseases have been identified, but access to skin biomarkers has largely been limited to invasive skin biopsies. |
| We hypothesized that chemically modified microprojections could provide a minimally invasive method to efficiently capture mRNA from the skin. |
| Using this platform, we have successfully and reproducibly extracted the transcriptome both from the skin of healthy individuals as well as skin lesions of psoriasis patients. |
| The dermal biomarker patch platform described herein could enable access to previously inaccessible datasets and enable novel study into the skin transcriptome. |
| Application of this platform could lead to high-precision diagnostics as well as the realization of precision medicine in dermatology. |
Introduction
Analysis of biomarkers has become a preferred method for early detection of disease, patient stratification, and monitoring efficacy of treatment [1]. Biomarkers can be used in clinical practice to identify risk for or diagnose a disease, stratify patients, assess disease severity or progression, predict prognosis, or guide treatment. In drug development, biomarkers may be used to help determine how a drug works in the body, to determine a biologically effective dose of a drug, to help assess whether a drug is safe or effective, and to help identify patients most likely to respond to a treatment, to predict who is at risk for relapse, or are least likely to suffer an adverse event when treated with a drug. Biomarkers can sometimes be used as part of the approval process for a drug or treatment, to inform regulatory decision-making. However, rapid and highly sensitive detection of changes in a biomarker is often technically impossible, or may require a cumbersome procedure involving multiple processing steps, necessitating large sample volumes and a prolonged diagnosis/prognosis timeline. The sample from a patient is often of a limited volume and not amenable to extensive processing or to procedures requiring multiple steps that extend the processing time.
Current methods for detecting molecular biomarkers or biological analytes of interest for diagnostic applications rely primarily on extraction of body fluid (e.g., blood, interstitial tissue fluid) from a patient. It is from this sample of fluid that specific biomarkers are assayed. More recent inventions describe the use of devices to facilitate the extraction of interstitial tissue fluid, including hollow microneedles [2], sonophoresis [3], and iontophoresis [4] technologies. However, these techniques do not directly sample clinically relevant biomarkers from the site of application, but rather, require further processing of body fluids. Other diagnostic methodologies that are not based on molecular assays (e.g., biopsies) cannot directly assess biomarker levels and can be subject to false-positive and false-negative results because of their inherently visual and subjective nature. However, for noncirculating biomarkers, this has often been the only diagnostic option to date.
Presently, the most common method for accessing molecules from the skin relies upon clinical biopsy of the site, followed by either histological examination of skin sections or homogenization of the biopsy sample with subsequent biomarker purification and downstream analysis; for example, for RNA-Seq-based studies, samples are homogenized and the mRNA purified, followed by library preparation and next-generation sequencing (NGS) protocols. Biopsy is an inherently invasive, scarring method that cannot be used to temporally follow a piece of skin throughout a study; once the sample is removed, the progression of the skin sample can no longer be monitored, thus providing only a single snapshot in molecular time. Furthermore, in studies where biopsy is not part of the clinical standard of care, it can be extremely difficult to have research subjects agree to have biopsies taken from their skin. It is also important to note that, from a diagnostic perspective, in many diseases, interpathologist concordance from biopsy samples is poor. Indeed, a 1996 study by Farmer et al. showed that 38% of cases had two or more discordant opinions when observed by eight different pathologists [5]. Additionally, in the case of skin cancer, Brochez et al. found that, on average, pathologists missed 13% of melanomas [6]. Finally, a 2008 work by Lodha et al. studying diagnostically challenging melanocytic neoplasms in the clinical setting showed a high level of disagreement among consultants in 25% of cases [7]. In essence, the primary technology used to examine the skin has changed little over the past 50 years.
Microneedles have recently become widely studied as a technology for transdermal delivery of drugs and/or biotherapeutics that would otherwise have poor penetration across the skin and, in particular, through the stratum corneum, the 15–20 µm tough outer layer of the epidermis. In general, microneedles are protrusions of 50–2000 µm in length, in varying shapes, that can be inserted into the skin and penetrate the epidermis and upper dermis without contacting the nerve endings and blood vessels that permeate the underlying dermis. Thus, in the context of drug delivery, microneedles have been touted as a pain-free method to deliver pharmaceuticals including small molecules and larger proteins.
Microneedles can be made of a wide variety of materials including silicon, stainless steel, glass, ceramic, and various polymers both synthetic and natural [8]. Most commonly, microneedle arrays are fabricated either from polymer or metal substrates. To make the former, techniques from the fabrication of microprocessors are applied. This technology has been widely applied in biological settings and other lab-on-a-chip devices and have been extensively reviewed [9, 10].
Given the relatively insensitive and cumbersome methodology currently utilized for skin research and skin disease diagnosis, it is clear that there is an urgent need for new technologies that allow for the sensitive measurement of skin-relevant biomarkers. In recent years, microneedle-based platforms have seen increasing use in transdermal diagnostics, particularly in the context of protein, metabolite, or drug detection from interstitial fluid [11–13]. However, these approaches have primarily relied upon colorimetry, immunoassays, and electrochemistry as a design. We have developed a dermal biomarker patch (DBP) that enables pain-free, minimally invasive, and targeted extraction of the transcriptome from the skin, without requiring a skin biopsy (Fig. 1). Herein, we describe the utility of these patches in extracting dermal transcriptomic information from human subjects.
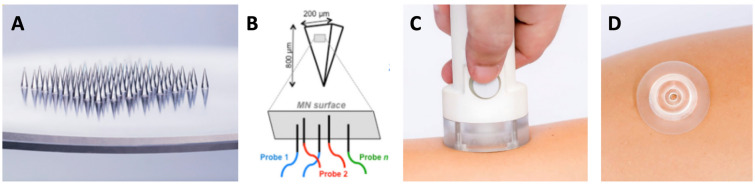
Fig. 1.
Mindera dermal biomarker patch. A Patches are approximately 2 cm in diameter with an array of 100 square pyramidal microneedles in the center of the patch arranged in a 5 × 5 mm2 square. Each pyramidal microneedle is 750 µm in length and 200 µm × 200 µm at the base. B Representation of modified microneedles with ssDNA probes bound to the surface; C application of dermal biomarker patch to the skin using spring-loaded applicator; D dermal biomarker patch applied to skin for sample collection
Methods
Patch Fabrication and Surface Modification
Custom-designed dermal patches (Accumold, Inc, Des Moines, IA) were chemically modified by first performing plasma treatment on the naked cyclo olefin polymer (COP, Zeonor 1020R) using a Surfx AtomFlo 500 instrument (Surfx Technologies, Redondo Beach, CA). The parameters used for plasma treatment were 120 W, 20 L/min argon, 0.20 L/min oxygen, and 60 °C CCM. Plasma treatment was conducted at 1 mm/s pass speed. After plasma treatment, chemical modification of patches was accomplished as follows: Fresh stock of 2 mM COMU (Sigma) intermediate was prepared in dimethylsulfoxide (DMSO), and N,N-diisopropylethylamine (Sigma) was then added to the reaction for 20 min. Activated COMU solution (50 µL) was added to the microneedle array and allowed to incubate at room temperature for 30 min. The dermal patch was then washed with acetone and dried under a stream of nitrogen. Oligo-dT (IDT 5′-/5ILINK12/iSp18TTTTTTTTTTTTTTTTTTTTTTTT-3′) was then diluted to 10 µM using 1X PBS. To the activated microneedle array, 75 µL oligo dT was added and allowed to incubate at room temperature overnight in a humidified chamber. The probe-modified dermal patch was washed with water and dried under a stream of nitrogen. Completed dermal patches were then sealed until use.
Fluorescence Monitoring of Modified Dermal Biomarker Patches
Modified dermal patches were examined by fluorescence microscopy to ensure consistent modification. For this, patches were modified adjacent to the microneedle array as follows. To the side of the array, 1 µL freshly made 2 mM COMU solution was added and then incubated for 30 min at room temperature. The patch was then washed with acetone and dried under a stream of nitrogen. Once dry, 1 µL oligo-dT (IDT 5′-/5ILINK12/iSp18TTTTTTTTTTTTTTTTTTTTTTTT-3′) was added to the same spot to couple the ssDNA probe to the patch surface. The patch was then incubated overnight at room temperature in a humidified chamber without shaking, then washed with water and dried under a stream of nitrogen. These modified spots were then treated with 20 µL 0.5 µM CY3-A24 oligonucleotide (Integrated DNA Technologies) and allowed to incubate at room temperature for 15 min. After this time, patches were washed with phosphate-buffered saline (PBS), dried under a stream of nitrogen, and imaged for fluorescence. The ratio of the fluorescence of the hybridized region versus the nonhybridized region was measured using ImageJ. Control experiments were performed where patches that did not have oligo-dT coupled, as well as those without olig-dT and without CY3-A24, were also imaged.
Human Subject Recruitment and Enrollment
All ex vivo skin samples were discarded skin obtained during the course of routine surgical procedures. All samples were deidentified, and no patient information was recorded. For in vivo experiments, four healthy subjects and three patients with active psoriasis were enrolled. For this study, approval was received for all ex vivo samples from the Institutional Review Board of the University of Rochester Medical Center, under a protocol with Sherrif Ibrahim as principal investigator. All in vivo sample acquisition was conducted under a protocol approved by the Integreview Institutional Review Board with Tobin J. Dickerson as principal investigator. All studies were performed in accordance with the Helsinki Declaration of 1964 and its later amendments. All subjects provided informed consent to participate in the study, and no identifying information was collected.
The study was not powered for statistical significance. For each normal subject, 2–4 DBPs were applied to the lower arm, while 1 DBP was applied to the lesional skin of each psoriasis patient. Additionally, 2-mm punch biopsies were taken from the lesional skin of psoriasis patients. After collection, samples were immediately preserved in RNALater (Thermo) and stored at 4 °C until use. For some extracted mRNA samples, multiple mRNA preamplifications were performed to assess technical reproducibility.
En Face Histology of Dermal Biomarker Patch Application
To assess the depth of penetration of dermal biomarker patches, ex vivo skin samples were sliced en face and resulting puncture sites quantified. Skin samples were cut to size, and the subcutaneous fat removed from the samples. The top of the skin was washed with ethanol and dried, after which time the DBP applicator was placed on the skin and the patch applied. The patch was left in place for 5 min, after which time the DBP was removed and the skin sample pressed between two pieces of corkboard to ensure that the skin remained flat during fixation. This was then submerged in 10% neutral buffered formalin for 24 h at room temperature. The fixed skin was then embedded in paraffin using standard methods (Histo-Tec, Hayward, CA) and 20 of 20-µm-thick slices were prepared for microscopic analysis. All slides were imaged, and the resulting images were imported into ImageJ and analyzed for microneedle penetration.
Dermal Biomarker Patch Application
To apply DBPs to the skin, a customized spring-loaded applicator was used. This applicator served to standardize the application pressure across subjects and across users (pressure applied ~ 120 N). The loaded applicator was placed against the skin, and the trigger pressed, applying the patch to the skin. The patch was then held in place against the skin for 5 min by a ring of medical tape. After this time, the patch was removed from the subject, immediately placed into storage buffer (LiCl, Triton X-100, Tris–EDTA), and stored at 4 °C until processing.
Dermal Biomarker Patch Processing
Dermal transcriptomes were processed within 72 h of collection from subjects. Samples were prepared by washing the applied DBP with chilled 1X PBS and then drying the patch under a stream of nitrogen. The dried DBP was then place on a heat block preset to 95 °C for 1 min, at which time, 50 µL PCR-grade water previously heated to 95 °C was then applied to the microneedle array for 1 min to elute the bound mRNA from the DBP. This eluted mRNA was then converted to cDNA using Takara SMART-Seq Single Cell kit according to the manufacturer’s instructions. Amplified cDNA samples were then stored at 4 °C until qPCR or NGS analysis.
Sequencing Procedures
Amplified cDNA was sequenced by a commercial vendor (Psomagen, Inc., Rockville, MD) according to standard procedures. Library preparation was accomplished using Illumina Nextera DNA Flex kits according to the manufacturer’s instructions. Prepared indexed libraries were then loaded onto a NovaSeq6000 S4 with read length of 150PE for sequencing of 40M reads per sample. During sequencing, the quality score (Q30) was maintained over 75%. Upon completion of runs, FASTQ file quality was checked with FASTQC and trimmed with the Trim_galore program. The trimmed FASTQ were aligned and mapped to human reference genome GRCh38 using the hisat2 program. The number of reads was counted for each Ensemble gene ID using the FeatureCounts program and Homo sapiens GRCH38.84.gtf. RNA expression analysis was further processed using the Bioconductor package edgeR. Genes were filtered using filterByExpr before logCPM (counts per million reads) were calculated. Plots were made using the ggplot2 and heatmap.2 functions in R. Percent of genes detected was determined using the ratio of genes detected (with > 0 count) over the total number of Ensemble genes.
Results
Polymer Modification Including QC Spot
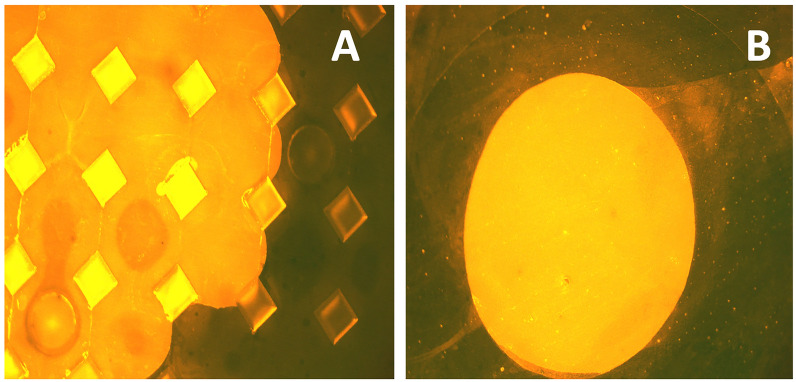
Polymer patches were chemically modified using atmospheric-pressure oxygen plasma to functionalize the surface for coupling with oligonucleotides [14]. This process was optimized to modify the surface without leading to visually apparent degradation of the pyramidal microneedles. Once functionalized, oligonucleotides were coupled to the surface using uronium salt chemistry to yield ssDNA-functionalized dermal biomarker patches that were then capable of binding to target mRNA. Assessment of the modification was performed by using the same chemistry on an adjacent portion of the back plate of the DBP. This spot was then treated with CY3-modified complementary ssDNA (CY3-A24), resulting in hybridization that could be measured by fluorescence microscopy (Fig. 2). A signal difference between modified and unmodified regions of the patch of at least threefold was achieved in all patches used in this study. Importantly, control experiments where no ssDNA chemical modification of the patch were performed, with no fluorescence being observed.
Fig. 2.
Fluorescence imaging of dermal biomarker patches. DBP were chemically modified adjacent to the microneedles with T24 oligonucleotides and then CY3-A24 hybridized for imaging. Strong fluorescence between the modified and unmodified regions can be seen and was quantified using ImageJ. A Fluorescence imaging of the DBP; B fluorescence imaging of spot on the DBP backplate used for quantitative assessment of chemical modification
En Face Histological Examination of Ex Vivo DBP Penetration
To examine the penetration of patches into human skin, deidentified excess skin explants were treated with modified DBPs. After application to the skin for 5 min, DBPs were removed, the skin fixed with formalin, and the skin embedded in paraffin for analysis. The resulting slides showed clear penetration of the microneedles into the skin to depths of ~ 350–400 µm. Additionally, from the size of the holes in each slice, an image of the microneedle was created that matches the pitch of the original device, validating that efficient penetration was achieved; examination of slices down to 300 µm showed no evidence of stratum corneum around the penetration site, as opposed to manual device application which showed ~ 15% of sites with stratum corneum present at depths of 300 µm.
In Vivo Transcriptome Extraction from Healthy Subjects
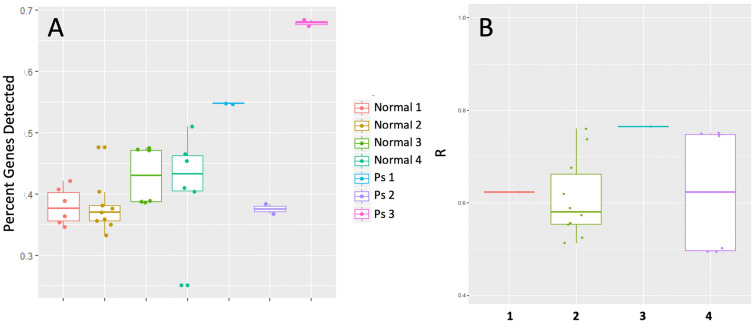
A total of 33 samples from seven subjects were sequenced for the assessment of technical and clinical feasibility (Fig. 3). RNA preamplification of all samples generated sufficient DNA quantity and quality for the preparation of sequencing libraries. The resulting DNA quantity ranged from 7.1 to 57.2 µg/µL, with an average concentration of 28 µg/µL. After sequencing, the percent of total genes detected was found to range from 25.1% to 68.3% of total genes (average 42.3%) across the 33 samples (Fig. 4A). Two of three psoriasis patients had the higher gene detection rates, which were ~ 54.7% and 68.8% respectively. The gene detection rates significantly correlated with DNA concentrations (R = 0.92, p < 0.001), suggesting that the amount of mRNA extracted from the skin onto the DBP was a major factor affecting the detection sensitivity. Importantly, unmodified DBPs that were not modified by ssDNA did not yield sufficient quantities of mRNA for sequencing library development.
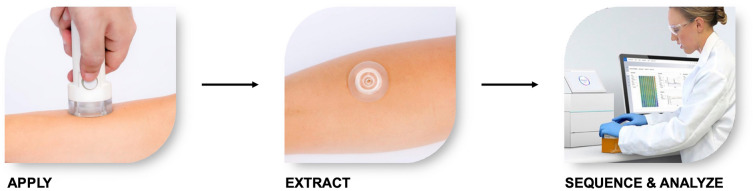
Fig. 3.
Dermal biomarker patch application, processing, and analysis. Briefly, patches are applied to skin using an applicator and allowed to remain on the skin for 5 min. After this time, the DBP is removed and the bound biomarkers eluted from the surface prior to next-generation sequencing and analysis
Fig. 4.
Analytical and clinical feasibility of Mindera dermal biomarker patch. A Percent of genes detected in DBP extractions from 33 samples; B biological reproducibility of gene intensity. Boxplot of correlation coefficients (R) of logCPM values from pairwise comparisons of different biological samples collected from each normal subject
To assess the biological reproducibility of the DBP platform, correlation coefficients (R) were calculated for each normal subject using logCPM values from the samples collected from different DBPs. R values ranged from 0.44 to 0.97, with an average of 0.69 across all samples (Fig. 4B).
Comparisons were also made between samples collected with the DBP and those collected via punch biopsy. In this case, samples collected with DBP were sequenced as before and then compared with tissue samples collected and processed using standard procedures with punch biopsy. Importantly, the correlation between punch biopsies and DBP collection was very good (Fig. 5), with excellent overlap in the number of genes detected by the two methods.
Fig. 5.
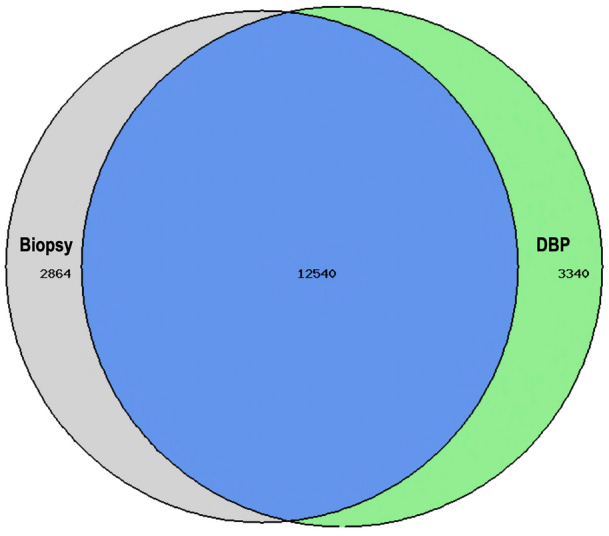
Overlap of genes detected in biopsy versus dermal biomarker patch collection. Of the total number of detected genes, 67% were detected in both DBP and biopsy, 18% were detected in DBP alone, and 15% were detected in biopsy alone
Discussion
Chemical modification of polymer surfaces for the extraction of biomarkers from in vivo skin has been successfully demonstrated using the described dermal biomarker patches. The covalently attached probes are able to bind specifically to mRNA and extract in sufficient quantity and quality for downstream next-generation sequencing. While direct measurement using fluorescence-based technologies are not possible at the mRNA quantities obtained, we have used amplified cDNA to assess the yield of genetic material captured from each patch. It is important to note that, unlike typical total RNA measurements, mRNA quality assessments by gel-electrophoresis-based methods cannot be performed as an RNA integrity number (RIN) is dependent upon detection of 28S and 18S rRNA peaks; since the DBP collects only mRNA, no RIN number can be calculated as there are no 28S or 18S rRNA peaks. Nonetheless, extraction of analyzable mRNA is only achieved with ssDNA-modified DBPs, demonstrating that the chemical modification is required.
With this information in hand, we next examined the ability of the DBP to extract molecular information from healthy skin and a representative disease phenotype, psoriatic lesions. Gratifyingly, meaningful quantities of mRNA were extracted from both skin types and the quantity of mRNA obtained was more than sufficient for the generation of next-generation sequencing libraries and subsequent sequencing. Furthermore, there was excellent overlap between the transcriptomes detected using DBP and punch biopsy.
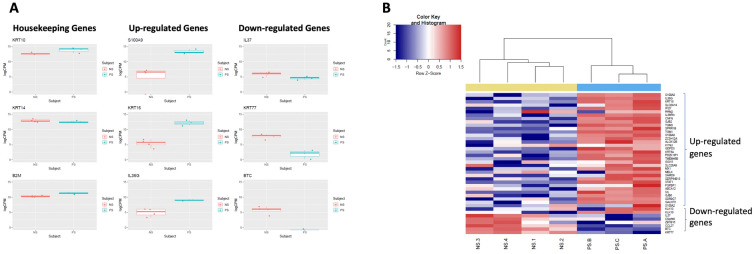
The two skin phenotypes examined in this study were compared to determine whether the DBP could detect transcriptomic differences between the two groups. As expected, many genes were not differentially expressed between the two groups, but there were a number of genes that showed good levels of up- or downregulation and were consistent with literature (Fig. 6A). Interestingly, genes such as S100A9, KDRT16, IL36G, and IL37 have not only been linked to differential expression in psoriatic lesions but also shown to correlate with patient response to biologic therapy [15–18]. To further validate the feasibility of DBPs in separating normal skin from psoriatic lesions, a list of 42 genes differential expressed in psoriasis were collected from literature, including 36 upregulated and 6 downregulated genes. As shown in Fig. 6B, these 42 genes were also consistently detected as upregulated or downregulated in DBP samples, confirming the feasibility of this platform for the extraction of mRNA and transcriptome profiling of diseased skin.
Fig. 6.
Differential expression of genes between normal skin and psoriatic lesions sampled with the DBP. A Expression levels of nine genes (three housekeeping genes, three up-regulated genes, and three downregulated genes in psoriasis) from seven subjects (four normal subjects and three psoriasis patients). B Heatmap of expression level (logCPM value) of 42 genes from seven subjects. All 42 genes showed consistent directional (up or down) regulation in the current dataset. NS, normal subjects; PS, psoriasis patients
Limitations
We note that this study was not designed to be statistically powered from a clinical perspective, but instead to show the ability of the dermal biomarker patch platform to extract the transcriptome from human skin. Future studies will be required to demonstrate statistically significant differences in clinical samples. Additionally, the differentially expressed transcriptome in psoriasis is a large number of genes. In this study, we did not aim to select a relevant subset of these genes for a specific purpose, but instead to show that the technology can reproducibly extract relevant biomarkers from the skin. Studies aimed at leveraging the data that can be acquired using these patches in psoriasis patients are left for future research.
Conclusions
Despite tremendous advances in our molecular understanding of the skin, visual observation and/or histology remains the standard for diagnosis and assessment. In many cases, such as inflammatory skin conditions, a routine method for collecting biomarker information from the skin is urgently needed to assist in disease diagnosis as well as to inform treatment paradigms. Relative to the dominant techniques used today (i.e., skin biopsy and pathology), the DBP solution brings a number of benefits including minimal invasiveness, high-precision molecular testing for improved sensitivity and specificity, and the generation of huge cost savings for healthcare systems as it can be cheaper than traditional pathology and lead to earlier and more accurate detection of disease. These data demonstrate the analytical and clinical feasibility of the DBP for skin transcriptome quantitation, enabling unprecedented study of the skin transcriptome and ready monitoring of the molecular “fingerprint” of the skin. In total, this platform has the potential to bring about widespread sampling and study of the skin, leading to high-precision diagnostics as well as novel tools that could realize the potential of precision medicine in dermatology.
Acknowledgements
We thank Dr. Changfu Wei for initial contributions to this project and Dr. Robert G. Eason for his contributions to our surface chemistry process development. We also gratefully thank the participation of the subjects of this study.
Funding
Support for this project and the journal’s Rapid Service fee was provided by Mindera Corporation.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
Project concept and design: Sherrif Ibrahim, Bradford Taft, Tahir Mahmood, Tobin Dickerson. Sample processing and wet laboratory procedures: Eric Andrade, Christian Abaya, Shreya Pramanick, Thejus Mannath, Katherine Hurley. Clinical procedures: Sherrif Ibrahim. Informatic analysis: Yipeng Wang, Drafting of the manuscript: Yipeng Wang, Byung-In Lee, Tobin Dickerson
Disclosures
Bradford J. Taft, Yipeng Wang, Byung-In Lee, Eric Andrade, Christian Abaya, Shreya Prmanick, Thejus Mannath, Katherine A. Hurley, and Tobin J. Dickerson are employees of Mindera Corporation. Sherrif F. Ibrahim receives research funding from Regeneron and funding for speaker’s bureaus services from Regeneron and Genentech. Sherrif F. Ibrahim, Yipeng Wang, Byung-In Lee, Tahir A. Mahmood, and Tobin J. Dickerson are shareholders of Mindera Corporation.
Compliance with Ethics Guidelines
For this study, approval was received for all ex vivo samples from the Institutional Review Board of the University of Rochester Medical Center, under a protocol with Sherrif Ibrahim as principal investigator. All in vivo sample acquisition was conducted under a protocol approved by the Integreview Institutional Review Board with Tobin J. Dickerson as principal investigator. All studies were performed in accordance with the Helsinki Declaration of 1964, and its later amendments. All subjects provided informed consent to participate in the study and no identifying information was collected.
Data Availability
The datasets generated during and/or analyzed during the current study are not publicly available as the data are proprietary to the sponsor of the study.
Contributor Information
Sherrif F. Ibrahim, Email: Sherrif_Ibrahim@URMC.Rochester.edu
Tobin J. Dickerson, Email: tdickerson@minderadx.com
References
- 1.Davies MA, Samuels Y. Analysis of the genome to personalize therapy for melanoma. Oncogene. 2010;29:5545. doi: 10.1038/onc.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prausnitz MR. Microneedles for transdermal drug delivery. Adv Drug Deliv Rev. 2004;56:581. doi: 10.1016/j.addr.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Tamada JA, Garg S, Jovanovic L, Pitzer KR, Fermi S, Potts RO. Cygnus research team. Noninvasive glucose monitoring: comprehensive clinical results. Cygnus Research Team. JAMA. 1999;282:1839. doi: 10.1001/jama.282.19.1839. [DOI] [PubMed] [Google Scholar]
- 4.Prausnitz MR, Langer R. Transdermal drug delivery. Nat Biotechnol. 2008;26:1261. doi: 10.1038/nbt.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farmer ER, Gonin R, Hanna MP. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum Pathol. 1996;27:528. doi: 10.1016/S0046-8177(96)90157-4. [DOI] [PubMed] [Google Scholar]
- 6.Brochez L, Verhaeghe E, Grosshans E, Haneke E, Piérard G, Ruiter D, Naeyaert JM. Inter-observer variation in the histopathological diagnosis of clinically suspicious pigmented skin lesions. J Pathol. 2002;196:459. doi: 10.1002/path.1061. [DOI] [PubMed] [Google Scholar]
- 7.Lodha S, Saggar S, Celebi JT, Silvers DN. Discordance in the histopathologic diagnosis of difficult melanocytic neoplasms in the clinical setting. J Cutan Pathol. 2008;35:349. doi: 10.1111/j.1600-0560.2007.00970.x. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly RF, Raj Singh TR, Woolfson AD. Microneedle-based drug delivery systems: microfabrication, drug delivery, and safety. Drug Deliv. 2010;17:187. doi: 10.3109/10717541003667798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao SL, Desai TA. Microfabricated drug delivery systems: from particles to pores. Adv Drug Deliv Rev. 2003;55:315. doi: 10.1016/S0169-409X(02)00227-2. [DOI] [PubMed] [Google Scholar]
- 10.Ingrole RSJ, Azizoglu E, Dul M, Birchall JC, Gill HS, Prausnitz MR. Trends of microneedle technology in the scientific literature, patents clinical trials and internet activity. Biomaterials. 2021;267:120491. doi: 10.1016/j.biomaterials.2020.120491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corrie SR, Fernando GJP, Crichton ML, Brunck MEG, Anderson CD, Kendall MAF. Surface-modified microprojection arrays for intradermal biomarker capture with low non-specific protein binding. Lab Chip. 2010;10:2655. doi: 10.1039/c0lc00068j. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Luan J, Seth A, Liu L, You M, Gupta P, Rathi P, Wang Y, Cao S, Jiang Q, Zhang X, Gupta R, Zhou Q, Morrissey JJ, Scheller EL, Rudra JS, Singamaneni S. Microneedle patch for the ultrasensitive quantification of protein biomarkers in interstitial fluid. Nat Biomed Eng. 2021;5:64. doi: 10.1038/s41551-020-00672-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu G-S, Kong Y, Wang T, Luo Y, Fan X, Xie X, Yang B-R, Wu MX. Microneedles for transdermal diagnostics: recent advances and new horizons. Biomaterials. 2020;232:119740. doi: 10.1016/j.biomaterials.2019.119740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gubala V, Le NCH, Gandhiraman RP, Coyle C, Daniels S, Williams DE. Functionalization of cyclo-olefin polymer substrates by plasma oxidation: Stable film containing carboxylic acid groups for capturing biorecognition elements. Colloids Surf B. 2010;81:544. doi: 10.1016/j.colsurfb.2010.07.055. [DOI] [PubMed] [Google Scholar]
- 15.Visvanathan S, Baum P, Vinisko R, Schmid R, Flack M, Lalovic B, Kleiner O, Fuentes-Duculan J, Garcet S, Davis JW, Grebe KM, Fine JS, Padula SJ, Krueger JG. Psoriatic skin molecular and histopathologic profiles after treatment with risankizumab versus ustekinumab. J Allergy Clin Immunol. 2019;143:2158. doi: 10.1016/j.jaci.2018.11.042. [DOI] [PubMed] [Google Scholar]
- 16.Krueger J, Clark JD, Suarez-Farinas M, Fuentes-Duculan J, Cueto I, Wang CQ, Tan H, Wolk R, Rottinghaus ST, Whitley MZ, Valdez H, von Schack D, P’Neil SP, Reddy PS, Tatulych S. Tofacitinib attenuates pathologic immune pathways in patients with psoriasis: a randomized phase 2 study. J Allergy Clin Immunol. 2016;137:1079. doi: 10.1016/j.jaci.2015.12.1318. [DOI] [PubMed] [Google Scholar]
- 17.Swindell WR, Remmer HA, Sarkar MK, Xing X, Barnes DH, Wolterink L, Voorhees JJ, Nair RP, Johnston A, Elder JT, Gudjonsson JE. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015;7:86. doi: 10.1186/s13073-015-0208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, Voorhees JJ, Abecasis GR, Elder JT. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol. 1829;2010:130. doi: 10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available as the data are proprietary to the sponsor of the study.