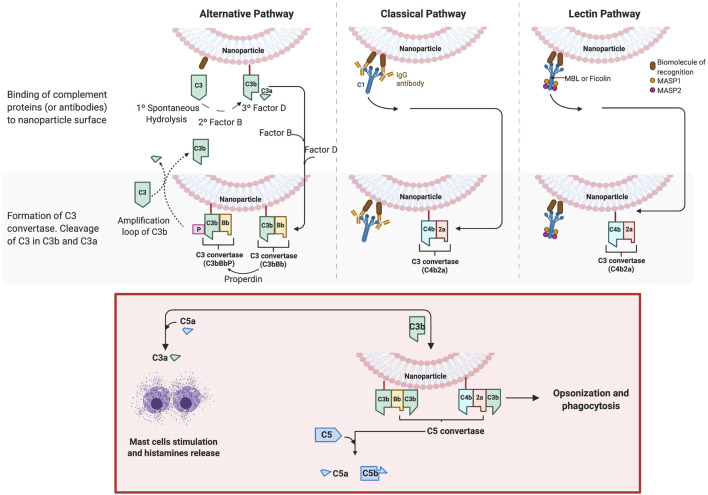
FIGURE 6.
Complement activation by nanoparticles. The activation of the complement cascade occurs via three different mechanisms: the classical, alternative and lectin pathways. The classical and lectin pathway is initiated by the recognition of patterns on the particle surface by soluble mediators in the blood. For the classical pathway, the mediators are antibodies, IgG or IgM. The Fc portion of these antibodies is recognized by the complement component C1, which includes C1q (recognition protein) associated with the proteases C1r and C1s. The binding leads the production of C3 convertase (C4b2a) (central panel). For the lectin pathway, the C3 convertase also is produced, but in this case, the recognition is carried out through other mediators, lectins, which recognizes sugars (mainly N-acetyl glucosamine and mannose). In humans, five different lectins that can initiate this pathway have been identified: MBL, M-, L-, and H-ficolins, and collectin 11 (CL11 or CL-K1), which are associated with other proteins, such as MASP-2 and MASP-1 (right panel). The synthetized C3 convertase cleaves the abundant serum protein C3 into C3a and C3b. Later, a larger fragment, C3b, is covalently attached to the surface. On the other hand, when the spontaneous hydrolysis of the thioester bond in the protein C3 takes place, the alternative pathway (left panel) is initiated. C3(H20) is thus produced, which binds to the factor B, another plasma protein, to form C3(H20)B. The latter is cleavage by the factor D, producing the convertase C3(H20)Bb, also called fluid-phase C3 convertase, which will cleave C3 in C3a and C3b. In turn, this forms C3b, which binds to factor B and then, in turn, is cleaved by factor D, forming C3bBb. C3bBb is stabilized by another complement protein, known as properdin (P), and forms the convertase C3bBbP, able to cleave C3 into C3a and C3b, establishing the amplification of more C3b that could bind on the particle (or pathogen) surface. C3(H2O) also can deposit (non-covalently) on surfaces and initiate C3 cleavage (Murphy et al., 2016; Moghimi and Simberg, 2017). The figure was prepared in BioRender.