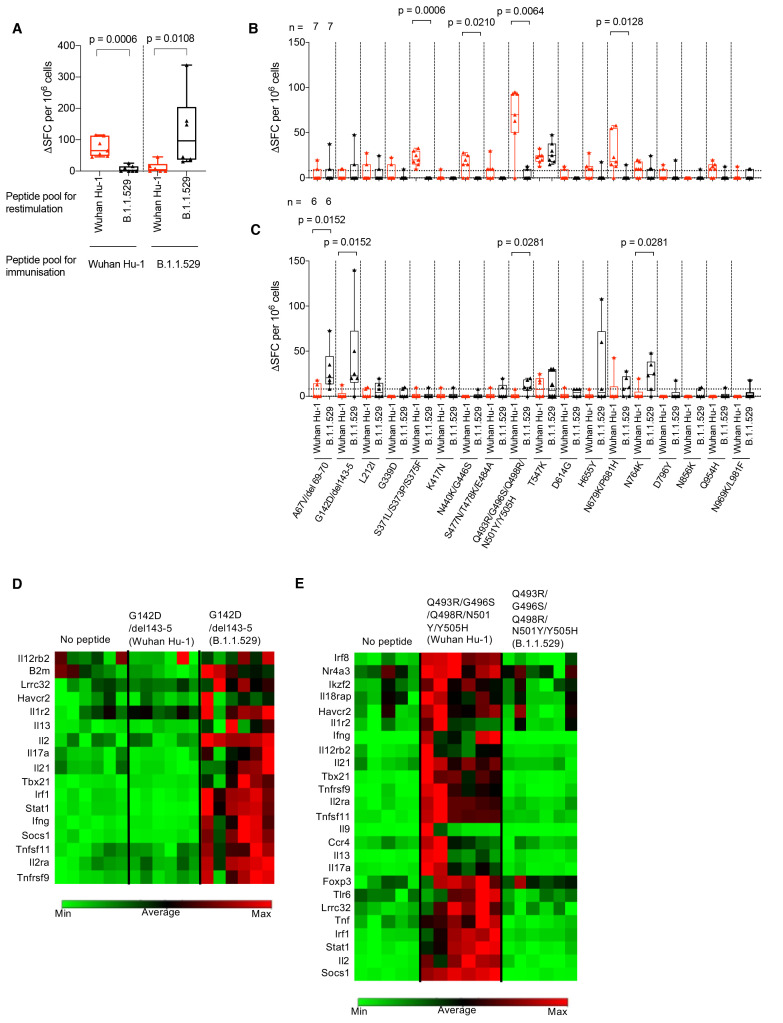
Fig. 3. B.1.1.529 (Omicron) spike mutations alter T cell recognition.
(A) HLAII transgenic mice carrying DRB1*0401 in the context of a homozygous knockout for murine H2-Aβ (7-8w) were immunized with either a B.1.1.529 (Omicron, n = 7) VOC pool of 18 peptides encompassing the Omicron sequence mutations or the ancestral Wuhan Hu-1 pool of peptides (n = 6) with the equivalent unmutated sequences. At d10 DLN cells were prepared from immunized mice and stimulated with either Wuhan Hu-1 (red) or B.1.1.529 (Omicron, black) peptide pools and T cell responses measured by IFNγ ELISpot. (B) IFNγ T cell responses were mapped against individual Wuhan Hu-1 (red) or B.1.1.529 (Omicron, black) peptides using DLN taken from Wuhan Hu-1 peptide pool (n = 7) or (C) B.1.1.529 (Omicron) peptide pool (n = 6) immunized mice. (D and E) Heatmaps showing relative gene expression of T cell activation markers in DLN cells taken from B.1.1.529 (Omicron) G142D/del143-5 peptide primed (D) (n = 6) or Wuhan Hu-1 Q493R/G496S/Q498R/N501Y/Y505H peptide primed (E) (n = 6) HLA-DRB1*04:01 transgenic mice. DLN cells were stimulated for 24h in vitro with 10 μg/ml Wuhan Hu-1 or B.1.1.529 (Omicron) variant peptide before RNA extraction. Genes shown in the heatmap are significantly up-regulated (p < 0.05) in Wuhan Hu-1 or B.1.1.529 (Omicron) variant peptide stimulated cells compared to no peptide control. Statistical tests were performed using Prism 9.0 or the Qiagen GeneGlobe data analysis tool for gene expression data. [(A) to (C)] Wilcoxon matched-pairs signed rank test, [(D) and (E)] Student’s t test. DLN, draining lymph node; SFC, spot forming cells; h, hours; VOC, variant of concern.