Fig. 3. The ubiquitin-proteasome system (UPP).
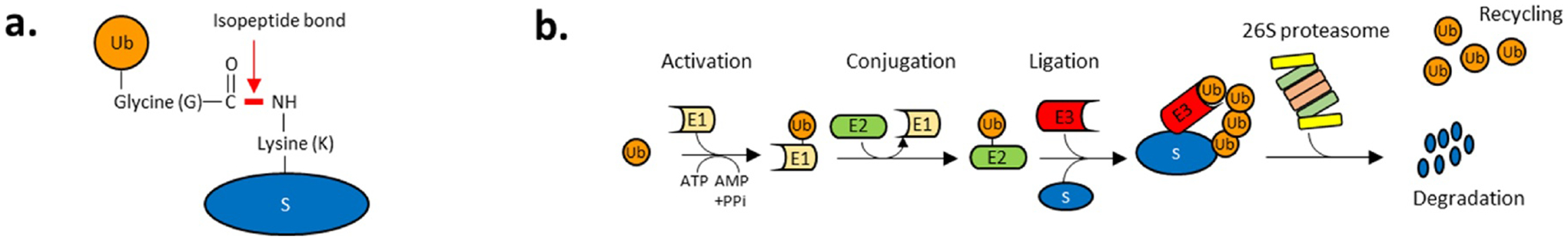
a. Ubiquitin (Ub) is a small protein (~76 amino acid) forming a covalent isopeptide bond between its c-terminal glycine carboxyl group and the NH2 group of a lysine residue on either substrate proteins or ubiquitin itself. b. Substrate ubiquitylation is achieved in three steps: substrate proteins are processed by ATP-activated enzymatic cascades with ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2) and ubiquitin ligases (E3). Ub is first activated by an E1 in an ATP-dependent manner. The E1 catalyzes the acyl-adenylation of the C-terminus of an ubiquitin molecule using ATP. The activated Ub is then conjugated to E1 through a thioester bond between the c-terminal carboxyl group of ubiquitin and the E1 cysteine sulfhydryl group (activation). E2 enzymes transfers the Ub molecule from E1 to the active site cysteine of an E2 via a trans(thio)esterification reaction (conjugation). E3 ligases allow the attachment of the Ub molecule from E2 to a substrate protein (S) by catalyzing the formation of an isopeptidyl linkage between a lysine residue of the substrate and the c-terminal glycine of the ubiquitin (ligation). Lysine 48-linked polyubiquitylation targets substrates to the 26S proteasome. The 19S regulatory particle of the proteasome docks the ubiquitylated substrate, unfolds its and deubiquitylates the substrate to recycle ubiquitin and transfer it to the 20S core particle where 3 proteolytic subunits degrade the substrate (see Fig. 4).
