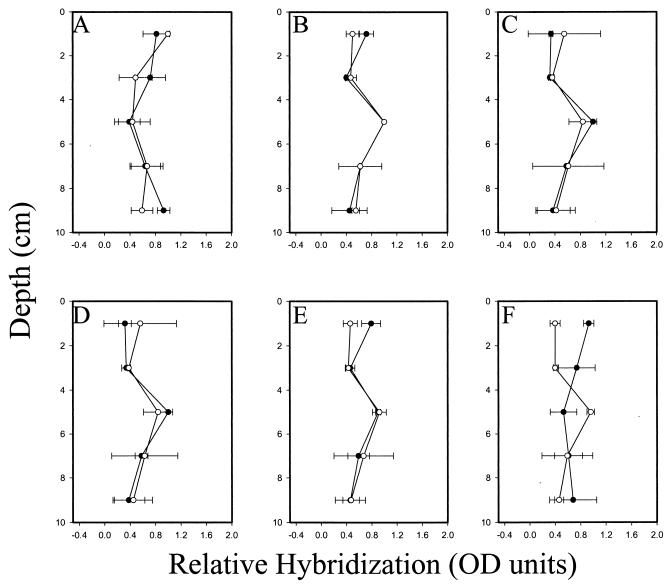
FIG. 4.
Relative depth distribution of total eubacteria and five groups of gram-negative mesophilic SRB in mercury- and PCB (Aroclor 1268)-contaminated salt marsh sediment equilibrated in the BERM mesocosm system for 6 months. Relative normalized hybridization is reported as a fraction of the strongest hybridization signal at any depth in a given core. Profiles of sulfate-reducing groups were normalized to the total eubacterial signal. Distribution was determined after whole-cell extraction (○) and direct RNA extraction (●). Depth profiles of total eubacteria (probe UNIV 342) (A), Desulfobacterium (probe SRB 221) (B), Desulfobacter (probe SRB 129) (C), Desulfovibrio (probe SRB 687) (D), Desulfococcus/Desulfosarcina/Desulfobotulus group (probe SRB 814) (E), and Desulfobulbus (probe SRB 660) (F) are shown. Error bars indicate standard deviation of the mean from four independent hybridizations. Depth distributions for each probe determined after whole-cell and RNA extractions were compared by two-way analysis of variance.