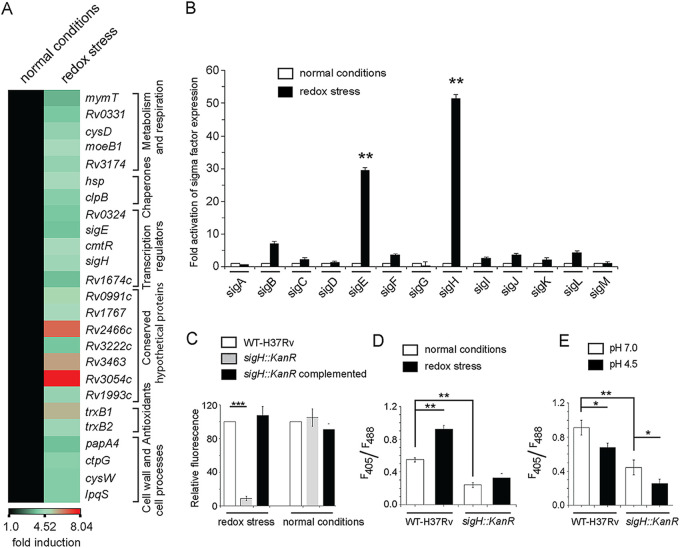
FIG 2.
Redox stress-inducible mycobacterial genes. (A) RNA-seq-derived heat map of WT H37Rv treated with 5 mM diamide versus WT H37Rv exposed to carryover DMSO. The data show a list of significantly induced genes (≥3.5-fold; P < 0.01), most of which belong to the SigH regulon. (B) To investigate redox-active sigma factors, expression of 13 mycobacterial sigma factor-encoding genes was compared by RT-qPCR using WT H37Rv grown in absence or presence of 5 mM diamide (compare empty and filled columns) as described in Materials and Methods. The results show average values from biological duplicates, each with two technical repeats (**, P ≤ 0.01). Nonsignificant difference is not indicated. (C) To compare the metabolic activity of the sigH::Kanr mutant with WT H37Rv, cells were grown in the presence of increasing concentrations of diamide, and alamarBlue assays were carried out as described in Materials and Methods. Bacterial metabolic activity under normal conditions (in the presence of carryover DMSO concentrations) and in the presence of 2.5 mM diamide was assessed by comparing the data from the mutant relative to those from the corresponding WT H37Rv cultures (considered 100%). (D and E) To examine if SigH contributes to mycothiol redox potential, we compared the intramycobacterial mycothiol redox states of the WT H37Rv and sigH::Kanr strains grown (D) under normal conditions versus in the presence of 5 mM diamide and (E) at pH 7.0 versus acidic pH (pH 4.5). Mycothiol redox state was determined as described in the legends to Fig. 1C and D.