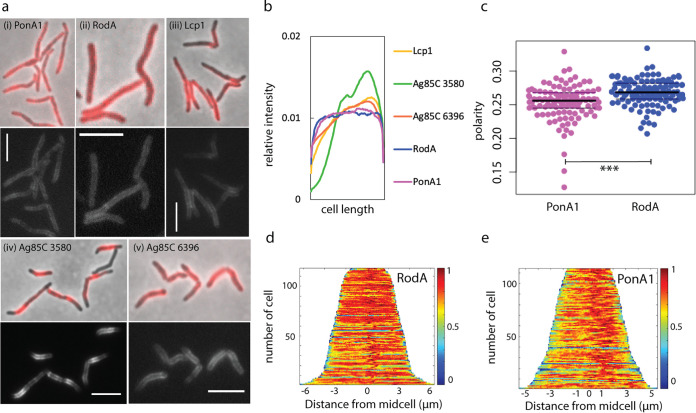
FIG 1.
RodA and PonA1 localize around the cell periphery under basal conditions. (a) Enzymes involved in peptidoglycan (i and ii), arabinogalactan (iii), and mycomembrane (iv and v) synthesis were fused to mRFP and imaged at an exposure of 5 s (i) or 4 s (ii to v). Scale bars, 5 μm. (b) Normalized mean fluorescence intensity profiles for mRFP-tagged enzymes. Fluorescence signals of each nonseptating cell were detected using Oufti and analyzed using MATLAB. Each cell was divided into 100 fragments, with each assigned an intensity value to normalize for cell length. Fragments for all cells were aligned to generate a mean intensity value for each fragment. These values were divided by the mean total cell fluorescence. 93 < n < 121. (c) Localization of RodA-mRFP and PonA1-mRFP was compared by calculation of the polarity ratio as a signal from both poles divided by the total cell fluorescence. t test, P < 0.001. (d and e) Normalized demographs. 112 < n < 118 (number of cells analyzed).