Figure 4.
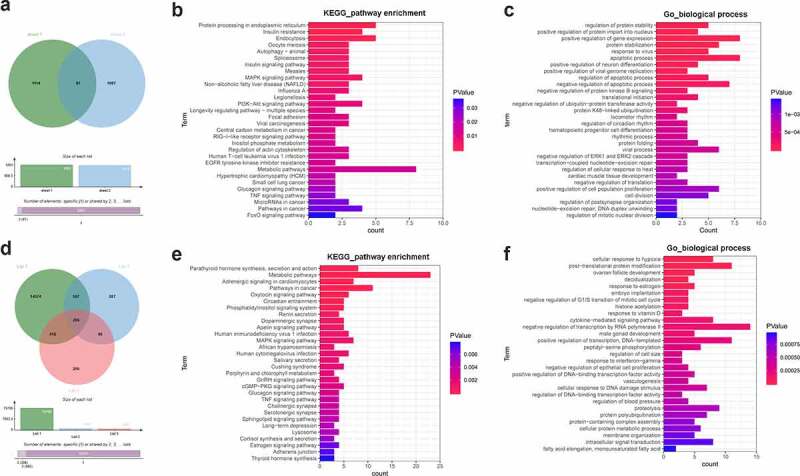
Identification of YTHDF3 and IGF2BP2 downstream regulatory pathways. (a) Identification of potential YTHDF3 targets. Sheet 1: m6a2target (http://m6a2target.canceromics.org/#/)-predicted protein-coding genes that could directly bind with YTHDF3. Sheet 2: Differentially expressed protein-coding genes after YTHDF3 knockdown in colorectal cancer cells. (b) The top 30 KEGG pathways enriched by the 87 potential YTHDF3 targets. (c) The top 30 biological processes enriched by the 87 potential YTHDF3 targets. (d) Identification of potential IGF2BP2 targets. List 1: Potential protein-coding genes with the probability to directly bind with IGF2BP2 were predicted by the Starbase database. List 2: Differentially expressed genes in IGF2BP2-depleted UMUC3 cells compared to the control group. List 3: Differentially expressed genes in IGF2BP2-depleted J82 cells versus the control group. (e) The top 30 KEGG pathways enriched by the 206 potential IGF2BP2 targets. (f) The top 30 biological processes enriched by the 206 possible IGF2BP2 targets.
