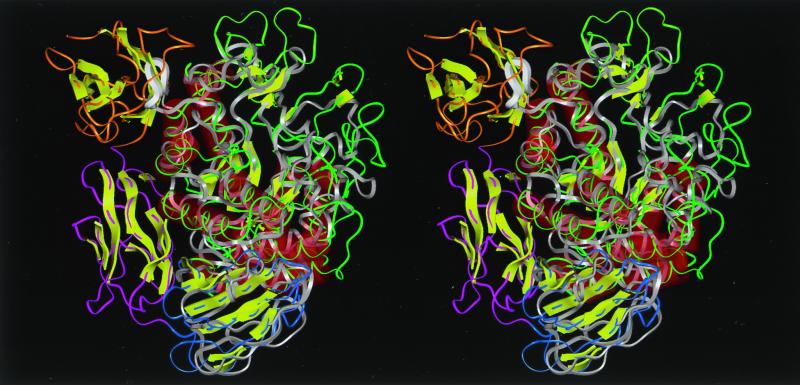
FIG. 4.
Stereoview of the predicted structure of Ba-S overlaid with the crystal structure of CGTase of B. circulans 251 (25) seen from the side of the (β/α)8-barrel. A structural comparison by aligning distance matrices was performed by the method of Holm and Sander (9). Molecular modeling of Ba-S was performed based on the three-dimensional structure of BSUA complexed with maltopentaose (5) using Discover-Insight II software (version 4.3; Molecular Simulation, Inc.) on an ONYX2 workstation (Silicon Graphics, Inc.). Residues 422 to 428 are not shown since the corresponding structure for BSUA could not be determined due to disorder (5). Ba-S is shown in white, and the loop between β4 and α4 of domain A (residues Pro180 to Ser187), which collides with a β-sheet structure (residues Asn635 to Pro643) in domain E of the CGTase, is shown as a thick ribbon. Green, yellow-green, blue, pink, and orange indicate domains A, B, C, D, and E of the CGTase, respectively. Red cylinders and yellow arrows represent α-helices and β-strands, respectively.