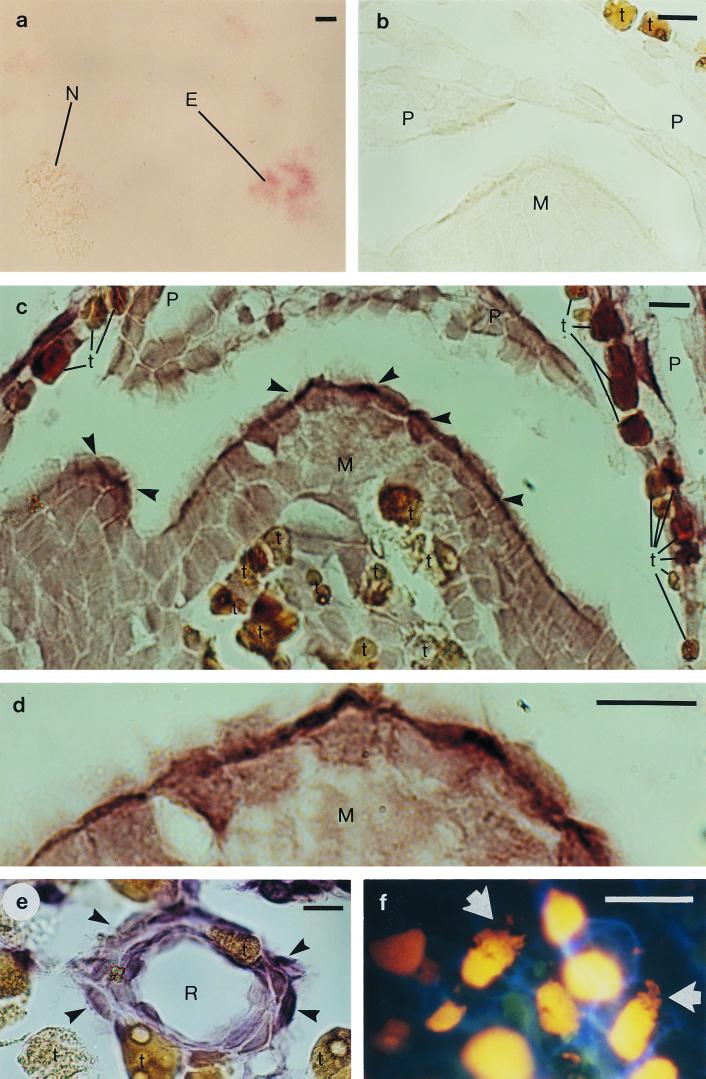
FIG. 1.
In situ hybridization of pine (Pinus sylvestris L.) bud tissue with digoxigenin-labeled oligonucleotides complementary to bacterial 16S rRNA sequences, and DNA staining of pine bud tissue. (a) Pine needle chloroplasts, mitochondria, and E. coli cells, hybridized with the eubacterial probe E11. The signal is detected in the E. coli cells (E) but not in the chloroplasts or mitochondria (N). (b) A negative control: apical meristem (M) and scale primordia (P), hybridized without a probe. (c) Apical meristem and scale primordia, hybridized with probe E11. The strongest hybridization signal was detected in the outermost cells of the meristem (arrowhead). For differentiation from positively hybridized cells, the naturally brown cells of the pine tissue, which contain tannins and other phenolic compounds, are marked with a t. (d) A magnification of the apical meristem from panel c. (e) A resin duct (R), hybridized with Methylobacterium-specific probe MB. The hybridization signal was detected in the cells surrounding the resin duct (arrowhead). (f) Ethidium bromide staining, revealing irregular structures (arrow) inside the cells, in addition to nuclei (yellow), surrounded by the cell walls (blue). Scale bar = 20 μm.