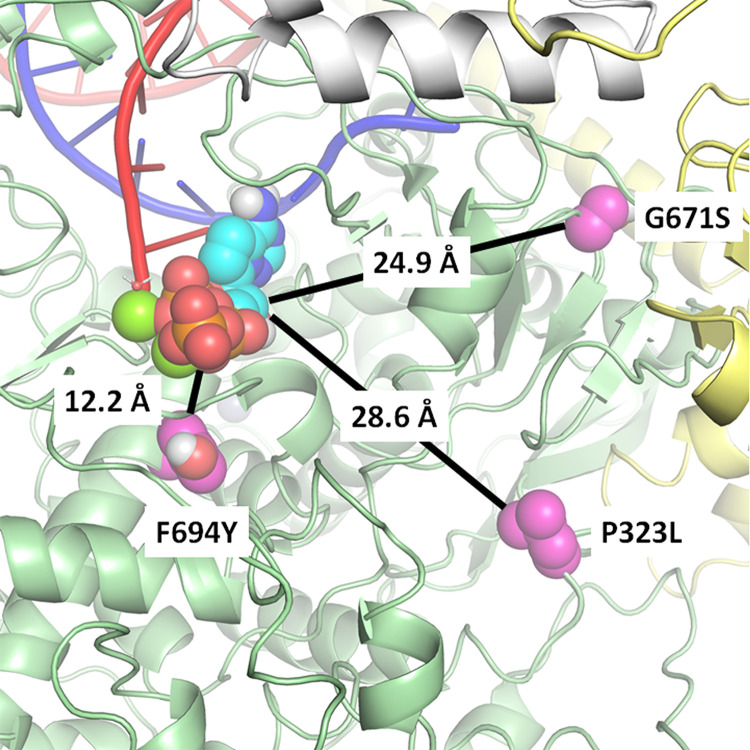
FIG 2.
Structural model of Nsp12 highlighting key amino acid substitutions in relation to the active site. Preincorporated remdesivir triphosphate (RDV-TP) was modeled into the cryo-EM structure of the polymerase complex (PDB no. 6XEZ) (37). Nsp12 is represented in green, with Nsp8 in yellow, and Nsp7 in white. The prevalent amino acid substitution P323L, seen in all variants, was measured to be 28.6 Å from RDV-TP (P323 Cα-RDV-TP C1′), whereas G671S, seen in the Delta variant, is at 24.9 Å. Of all the amino acid substitutions reported here, F694Y, seen at low frequency in Delta and Omicron, comes closest to the active site, at 12.2 Å. A computational analysis suggests that the substitutions have no meaningful impact on RDV-TP binding affinity.