Abstract
In a recent publication (S. M. Sievert, T. Brinkhoff, G. Muyzer, W. Ziebis, and J. Kuever, Appl. Environ. Microbiol. 65:3834–3842, 1999) we described spatiotemporal changes in the bacterial community structure at a shallow-water hydrothermal vent in the Aegean Sea near the isle of Milos (Greece). Here we describe identification and phylogenetic analysis of the predominant bacterial populations at the vent site and their distribution at the vent site as determined by sequencing of DNA molecules (bands) excised from denaturing gradient gels. A total of 36 bands could be sequenced, and there were representatives of eight major lineages of the domain Bacteria. Cytophaga-Flavobacterium and Acidobacterium were the most frequently retrieved bacterial groups. Less than 33% of the sequences exhibited 90% or more identity with cultivated organisms. The predominance of putative heterotrophic populations in the sequences retrieved is explained by the input of allochthonous organic matter at the vent site.
Information about the microbial community structure of hydrothermal vent systems is necessary in order to gain a more thorough understanding of the functioning of these unique ecosystems and their impact on the surrounding environment. Vent-associated microorganisms are the basis of the food webs at such localities and may also be involved in microbially mediated transformation and precipitation of elements (12, 14). Selective enrichment cultivation is not considered a suitable tool for characterizing microbial communities (2, 19, 24, 36), and in several studies researchers have used methods based on analysis of 16S rRNA sequences to study the bacterial communities at deep-sea vent sites (9, 17, 18, 20, 25). These studies demonstrated that only a few specialized bacterial populations dominated the microbial communities under the extreme physicochemical conditions found at the vent sites examined. By using denaturing gradient gel electrophoresis (DGGE), Muyzer et al. (20) identified four phylotypes in samples taken from two vent sites on the Mid-Atlantic Ridge (MAR). Two of these phylotypes were closely related to sulfur-oxidizing Thiomicrospira spp. which were frequently isolated at a variety of vent sites, including the MAR (13, 39). Polz and Cavanaugh (25) found that at another MAR vent site the putative sulfur-oxidizing epibiont of a shrimp dominated the microbial community. At a hydrothermal vent system located on Loihi Seamount, Hawaii, a midplate volcano, one of the two operational taxonomic units that dominated the fairly diverse community was affiliated with the sulfur-oxidizing bacterium Thiovolum sp. (18). These results substantiated the earlier assumption that chemolitho(auto)trophy that depends on reduced sulfur compounds is an important process at vent sites (12, 14).
We have used a shallow submarine hydrothermal vent in the Aegean Sea near the island of Milos (Greece) to investigate the relationship between changes in physicochemical parameters and bacterial population distributions by using DGGE of PCR-amplified 16S rRNA gene fragments (31). In this paper we describe identification of the dominant 16S rRNA-defined bacterial populations along a transect from the center of the vent out into the surrounding sediment. Bands were excised from DGGE gels and sequenced in order to obtain information about the phylogenetic affiliations of the dominant populations and to make inferences about the trophic structure of the microbial communities at the vent site.
The study site was a solitary gaseous hydrothermal vent located in 8 m of water in Palaeochori Bay (24°31.220′E, 36°40.391′N). Sea grass beds consisting of Cymodocea nodosa (depth range, 6 to 20 m) and Posidonia oceanica (depth range, 10 to 40 m) were present in the bay (1). A more detailed site description, including physicochemical parameters, has been published previously (31). The various research projects being conducted in Palaeochori Bay have been summarized by Dando et al. (6). Sediment cores were taken with polycarbonate tubes by scuba divers along a transect from the center of the almost circular vent out into the surrounding area at locations 10, 123, 165, and 235 cm from the vent center in June 1996 and at locations 30, 117, and 200 cm from the vent center in September 1996. At a distance of 117 cm two cores [cores 117 (I) and 117 (II)] were taken 1 week apart. Each sediment core was immediately subsampled by slicing the extruded sediment as described previously (31).
DNA extraction from subsamples obtained from sliced sediment cores and PCR amplification were performed as described previously (31). Amplification products were first analyzed on agarose gels before further characterization by DGGE analysis or DNA sequencing.
DGGE was performed as described by Sievert et al. (31). Selected DGGE bands were excised from the DGGE gels, reamplified by PCR with primers GM5F and 907R, and reelectrophoresed on a DGGE gel to verify the purity of each band and its position relative to the position of the original band, as described previously (7). Before the PCR products were sequenced, they were purified by using a Qiaquick Spin PCR purification kit (Qiagen Inc., Chatsworth, Calif.). A Taq Dyedeoxy terminator cycle sequencing kit (Applied Biosystems, Foster City, Calif.) was used to sequence the 16S ribosomal DNA fragments with primers GM5F and 907R. The sequence reaction mixtures were electrophoresed with an Applied Biosystems model 373S DNA sequencer.
Sequences were added to the 16S rRNA sequence database of the Technical University of Munich (Munich, Germany) by using the ARB software program package (http://www.mikro.biologie.tu-muenchen.de). Sequences were first aligned automatically by using ARB_ALIGN and then were checked by eye and corrected manually. Only sequences that were at least 90% complete were used for tree construction. Partial sequences obtained from the DGGE analysis were inserted into the reconstructed tree by applying the parsimony criteria without allowing for changes in overall tree topology. Tree topologies were further evaluated by performing maximum-parsimony, neighbor-joining, and maximum-likelihood analyses.
Number of unique populations.
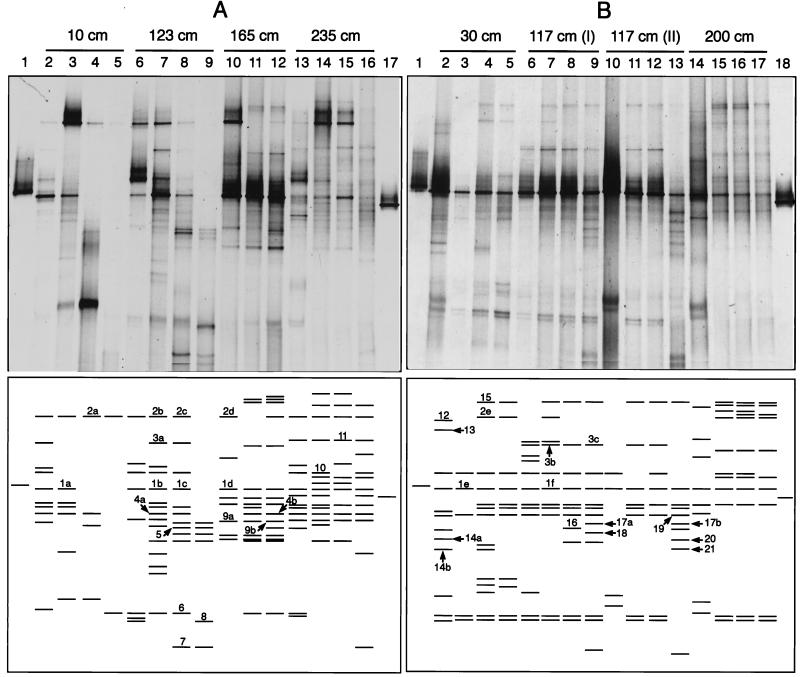
Figure 1 shows two DGGE gels that were prepared with samples collected in June 1996 (Fig. 1A) and September 1996 (Fig. 1B). The lower panels show all of the bands that could be visualized by DGGE. Altogether, we obtained 51 unique bands (i.e., bands with distinct electrophoretic mobilities) for the June samples (Fig. 1A) and 44 unique bands for the September samples (Fig. 1B). It is likely that these numbers of unique bands underestimated the actual diversity, since bands that have the same electrophoretic mobility can contain different sequences. However, in all cases in which we sequenced bands that had the same electrophoretic mobility, we found that the sequences were nearly identical (see below). Altogether, about 80 bands were excised from both gels, and 36 of these bands resulted in unambiguous sequences that were used for phylogenetic analysis. The other excised bands either resisted reamplification by PCR (36%) or yielded ambiguous sequences (64%). This could have been due to the presence of more than one sequence in a particular band (28).
FIG. 1.
DGGE analysis of 16S ribosomal DNA fragments obtained after PCR amplification with bacterial primers GM5F-GC-clamp and 907R of genomic DNA from environmental samples and standards with known melting behavior in June 1996 (A) and September 1996 (B). In the upper panels the actual DGGE gels are shown, whereas the lower panels are sketches that show the bands that were identified on each DGGE gel. The band numbers are the numbers for excised and sequenced bands, which are discussed in the text. The environmental samples were taken at specific locations along a transect from the vent center towards the surrounding area. The two cores obtained at a distance of 117 cm in September were taken 1 week apart. During the first sampling [core 117 (I)] a white precipitate was present on the sediment surface, whereas during the second sampling [core 117 (II)] there was no precipitate. No PCR product was obtained from the overlying water at a distance of 165 cm. (A) DGGE patterns for the samples taken in June 1996. Lanes 1 and 17, standards; lanes 2 to 5, samples taken 10 cm from the center of the vent (sediment depth shown in parentheses) (lane 2, surface; lane 3, 0 to 10 mm; lane 4, 10 to 20 mm; lane 5, 20 to 30 mm); lanes 6 to 9, samples taken 123 cm from the center of the vent (lane 6, surface; lane 7, 0 to 10 mm; lane 8, 10 to 20 mm; lane 9, 20 to 30 mm); lanes 10 to 12, samples taken 165 cm from the center of the vent (lane 10, 0 to 10 mm; lane 11, 10 to 20 mm; lane 12, 20 to 30 mm); lanes 13 to 16, samples taken 235 cm from the center of the vent (lane 13, surface; lane 14, 0 to 10 mm; lane 15, 10 to 20 mm; lane 16, 20 to 30 mm). (B) DGGE patterns for the samples taken in September 1996. Lanes 1 and 18, standards; lanes 2 to 5, samples taken 30 cm from the center of the vent (sediment depth shown in parentheses) (lane 2, surface; lane 3, 0 to 5 mm; lane 4, 8 to 13 mm; lane 5, 16 to 26 mm); lanes 6 to 9, samples taken 117 cm from the center of the vent (lane 6, surface; lane 7, 0 to 5 mm; lane 8, 8 to 13 mm; lane 9, 16 to 26 mm); lanes 10 to 13, samples taken 117 cm from the center of the vent (lane 10, surface; lane 11, 0 to 5 mm; lane 12, 8 to 13 mm; lane 13, 16 to 26 mm); lanes 14 to 17, samples taken 200 cm from the center of the vent (lane 14, surface; lane 15, 0 to 5 mm; lane 16, 8 to 13 mm; lane 17, 16 to 26 mm). Parts of this figure were reproduced from reference 31 with permission from the publisher.
Phylogenetic affiliation and spatial distribution of dominant populations.
Table 1 shows the sequences analyzed, the locations along the transect where they were obtained, their phylogenetic positions, and their putative physiologies inferred from the physiologies of the most closely related cultivated organisms. The distribution of the bands that were sequenced shows that the sequences represented bacterial populations that were present in different zones and at different sediment depths, as well as populations obtained at different sampling times (Fig. 1). However, fewer sequences were successfully retrieved from the outer zones at both sampling times. This might have been related to the greater complexity of the DGGE profiles in these regions (31), which resulted in a higher probability that particular bands contained more than one sequence (22). Our phylogenetic analysis of the bands revealed wide diversity within the domain Bacteria. Similar findings have been obtained for a variety of environments, including deep-sea hydrothermal vents (18) and terrestrial hot springs (11). The actual diversity might have been even higher because we sequenced only 28% of the unique bands obtained from the vent site examined, DGGE detects only dominant populations, and it is possible that bacteria specific to this habitat may not have contained the signature sites necessary for efficient amplification with the bacterial primers used.
TABLE 1.
Summary of the 16S rRNA sequences derived from excised DGGE bands shown in Fig. 1
| Band | Origin
|
Putative divisiona | Database match (≥90% identity) | % Identity | Group | Inferred metabolism | |
|---|---|---|---|---|---|---|---|
| Distance (cm) | Sediment depth (mm) | ||||||
| ML-1a | 10 | 0–10 | CFB | Deep-sea hydrothermal vent clone VC2.1 Bac22 | 96 | Cytophaga spp. | Organotrophy? |
| ML-1b | 123 | 0–10 | |||||
| ML-1c | 123 | 10–20 | |||||
| ML-1d | 165 | 0–10 | |||||
| ML-1e | 30 | 0–5 | |||||
| ML-1f | 117 (I)c | 8–13 | |||||
| ML-2a | 10 | 10–20 | Cyanobacteria and chloroplasts | Chloroplast (Odontella sinensis) | 98 | Bacillariophyta | Photosynthesis |
| ML-2b | 123 | 10–20 | |||||
| ML-2c | 123 | 20–30 | |||||
| ML-2d | 165 | 0–10 | |||||
| ML-2e | 30 | 8–13 | |||||
| ML-3a | 123 | 0–10 | CFB | Cytophaga spp. | Organotrophy? | ||
| ML-3b | 117 (I) | 0–5 | |||||
| ML-3c | 117 (I) | 16–26 | |||||
| ML-4a | 123 | 0–10 | Spirochetes | Organotrophy? | |||
| ML-4b | 165 | 20–30 | |||||
| ML-5 | 123 | 0–10 | Proteobacteria (δ) | Desulfobacteriaceae | Sulfate reduction? | ||
| ML-6 | 123 | 10–20 | Synergists | ||||
| ML-7 | 123 | 10–20 | Candidate division OP8 | ||||
| ML-8b | 123 | 20–30 | Candidate division OP9 | Clone OPB 46 | 94 | ||
| ML-9a | 165 | 0–10 | Acidobacterium | ||||
| ML-9b | 165 | 20–30 | |||||
| ML-10 | 235 | 0–10 | Proteobacteria (γ) | Thiomicrospira strain Milos T-2 | 99 | Thiomicrospira | Chemolithoautotrophic sulfur oxidation |
| ML-11 | 235 | 10–20 | CFB | Cytophaga fermentans | 91 | Cytophaga spp. | Organotrophy? |
| ML-12 | 30 | Surface | Cyanobacteria and chloroplasts | Chloroplast (Emiliana huxleyi) | 96 | Haptophyceae | Photosynthesis |
| ML-13 | 30 | Surface | Cyanobacteria and chloroplasts | Chloroplast (Emiliana huxleyi) | 97 | Haptophyceae | Photosynthesis |
| ML-14ab | 30 | Surface | Low-G+C gram-positive bacteria | Caldicellulosiruptor lactoaceticus | 97 | Caldicellulosiruptor | Organotrophy, fermentative degradation of polysaccharides |
| ML-14bb | 30 | Surface | Low-G+C gram-positive bacteria | Caldicellulosiruptor lactoaceticus | 99 | Caldicellulosiruptor | |
| ML-15 | 30 | 8–13 | CFB | Flexibacter | Organotrophy? | ||
| ML-16 | 117 (I) | 8–13 | Acidobacterium | ||||
| ML-17a | 117 (I) | 16–26 | Flexistipes | ||||
| ML-17b | 117 (II) | 16–26 | |||||
| ML-18 | 117 (II) | 16–26 | Acidobacterium | ||||
| ML-19 | 117 (II) | 16–26 | Proteobacteria (δ) | Desulfobacteriaceae | Sulfate reduction? | ||
| ML-20 | 117 (II) | 16–26 | Acidobacterium | ||||
| ML-21 | 117 (II) | 16–26 | Acidobacterium | ||||
CFB, Cytophaga-Flavobacterium-Bacteroides phylum.
Inferred thermophily.
117 (I) and 117 (II), samples obtained 1 week apart in September 1996.
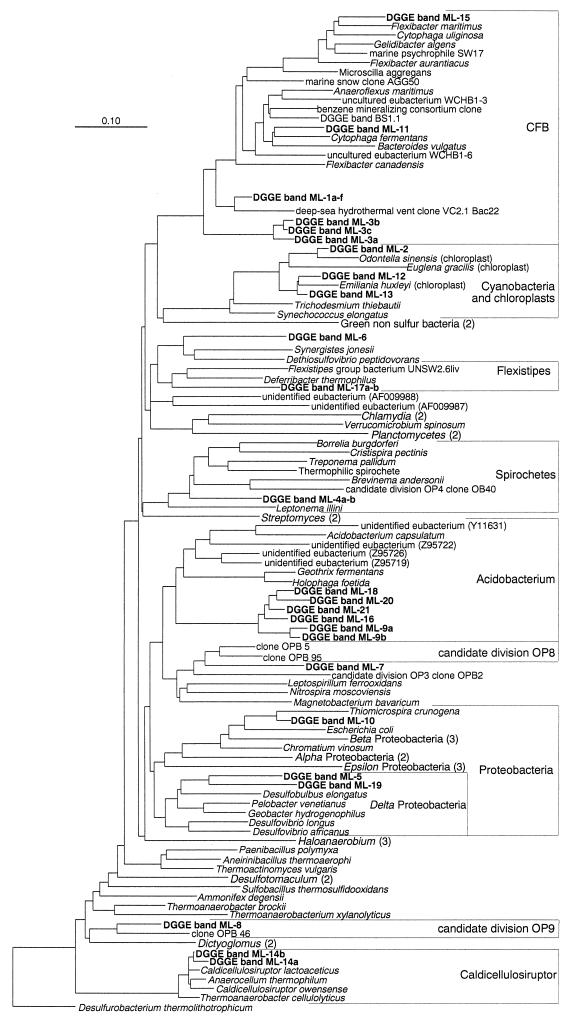
Eleven sequences were affiliated with the Cytophaga-Flavobacterium (CF) cluster of the Cytophaga-Flavobacterium-Bacteroides phylum, and thus, members of this cluster were the most prominent phylotypes detected. Of these 11 sequences, 6 came from the dominant band (band ML-1a through band ML-1f). All of these sequences were identical or at least extremely closely related (>99% identity). Following the suggestions of Fox et al. (8), we grouped all of these sequences into one “rRNA superspecies”; only one representative, band ML-1, is shown in Fig. 2. If it is assumed that the relative intensities of bands in DGGE gels are correlated with the actual sizes of the corresponding populations (21, 22), it can be inferred that the population corresponding to band ML-1 is a predominant member of the microbial community at our vent site, particularly in zones more strongly affected by the hydrothermal fluid. The fact that this population was most closely related to a clone (96% sequence identity) (Fig. 2) that was obtained from an in situ growth chamber deployed at a deep-sea hydrothermal vent (DDBJ, EMBL, and GenBank accession no. AF068798) provides support for the hypothesis that it is an indigenous and probably thermophilic population. The ML-3a-c sequences formed another group in the CF cluster (Fig. 2). The two sequences obtained from September samples were almost identical (>99% identity), and both exhibited 98% identity to the sequence obtained from June samples. These sequences had no close relatives in the database (levels of identity, <85%), and thus, their exact phylogenetic position in the CF cluster remains unclear. Two other sequences in the CF cluster, the ML-11 and ML-16 sequences, exhibited 92% identity to the Cytophaga fermentans sequence and 88% identity to the Flexibacter maritimus sequence, respectively (Fig. 2). Populations related to the CF cluster have also been reported to be important in other marine sediments, including North Sea (15) and Black Sea (28) sediments. Inferring the physiology of these populations from their phylogeny is difficult, but since all cultured members of the CF cluster are heterotrophic and many members degrade polymers (27), it seems likely that these sequences originated from populations that have similar metabolism. However, it is possible that some sequences represent lineages with new metabolic capabilities.
FIG. 2.
Evolutionary tree showing the phylogenetic affiliations of 16S rRNA sequences obtained from bands excised from the DGGE gels shown in Fig. 1. The tree was constructed with the software package ARB based on full-length sequences by using parsimony analysis. The partial ca. 550-bp sequences (boldface type) were inserted into the tree by using maximum-parsimony criteria without affecting the initial tree topology by using a special tool implemented in ARB. The sequence of DGGE band ML-10 has been determined previously by Brinkhoff et al. (5). The numbers in parentheses are the numbers of different sequences used to calculate the phylogenetic positions or accession numbers. Bar = 10% sequence divergence. CFB, Cytophaga-Flavobacterium-Bacteroides phylum.
Like the ML-1 sequences, the sequences that were derived from ML-2 in different profiles (i.e., ML-2a through ML-2e) were identical, and only one representative sequence of this rRNA superspecies is shown in Fig. 2. These sequences were affiliated with chloroplasts of diatoms, and the most closely related full-length sequence was the chloroplast sequence of the marine diatom Odontella sinensis (level of sequence identity, 98%) (Fig. 2). Based on the 415-nucleotide overlap among partial band ML-2, Navicula salinicola (23), and Amphora delicatissima (23) sequences, the band ML-2 sequence formed a cluster with these diatoms and was most closely related to N. salinicola (data not shown). The population corresponding to band ML-2 was also present at both times. However, while band ML-2 was clearly stronger in the upper layers and represented an important component of the microbial community in June, the importance of the population seemed to be minor in September. The population corresponding to the ML-2 sequence could in principle have originated from three sources: (i) sedimentation of planktonic species from the water column, (ii) in situ growth of benthic species on the sediment surface, and (iii) epiphytic species that were transported to the hydrothermal vent. We could eliminate the first possibility because the closest relatives of the sequence were species belonging to the pennate genera Navicula and Amphora and the members of both of these genera grow only on surfaces (29). The presence of the genus Amphora and the presence of the species N. salina were microscopically verified for samples obtained from different zones along the transect (A. Economou-Amilli, personal communication), and we also found pennate diatoms of unknown affiliation thriving in the white precipitate and at the center of the vent during microscopic inspection on site (S. M. Sievert and G. Kuever, unpublished data). It is interesting that these populations seemed to be active near the center of the vent, where the in situ temperature at the sediment surface was between 30 and 40°C. Most diatoms cannot grow at these temperatures, although the closest relatives of the organisms corresponding to band ML-2 originating from hypersaline mats in Guerrero Negro, Baja California (Mexico), were able to grow at 37°C (F. Garcia-Pichel, personal communication). Thus, we suggest that the diatoms corresponding to band ML-2 contributed to production of autochthonous organic matter at the vent site by carrying out photosynthesis; photosynthesis has been measured at similar vent sites in Palaeochori Bay (37). The closest relatives of the chloroplastic ML-12 and ML-13 sequences were members of the Haptophyceae (97% identity to the chloroplast 16S rRNA sequences of Emiliana huxleyi and Orchrosphaera neapolitana) (Fig. 2). Both E. huxleyi and O. neapolitana are planktonic marine algae, and thus, the sequences probably originated from planktonic species. However, unknown benthic forms might exist.
We found three sequences which were affiliated with members of the class Proteobacteria (Fig. 2). One sequence (ML-10) was almost identical to the sequence of Thiomicrospira sp. strain Milos T-2, a member of the gamma subclass of the Proteobacteria, which was isolated from samples obtained at the same vent site (5). The population corresponding to band ML-10 was present at both sampling times, and by performing hybridization analysis with a specific probe, we demonstrated that all bands that migrated to the same position on the gel were produced by sulfur-oxidizing Thiomicrospira populations (5). The finding that Thiomicrospira spp. constitute an important component of the microbial community suggests that besides photosynthesis, chemosynthesis contributes to primary production at the vent site studied. Isolation of other chemolithoautotrophic sulfur-oxidizing bacteria from similar or even higher dilutions provided further support for the hypothesis that chemosynthesis is important in this habitat and revealed a high level of diversity for the sulfur-oxidizing bacteria (30). The band ML-5 and ML-19 sequences were loosely affiliated with members of the delta subclass of the Proteobacteria (Fig. 2). Detection of these bands could be an indication that sulfate reduction is an important electron acceptor for terminal oxidation of organic matter in this habitat. Sulfate reducers were present in this environment and were most abundant 235 cm from the center of the vent (31). This was also the region in which the highest sulfate reduction rates were observed (W. Ziebis, V. Brüchert, S. Forster, and B. B. Jørgensen, unpublished data). A bacterium that was most closely related to Desulfobacter halotolerans (4) (97% 16S rRNA identity) was isolated from the highest dilution of a most probable number (MPN) series (30). Desulfobacter spp. have a narrow substrate spectrum and are thought to be specialists for degradation of acetate (38). Thus, their relatively high numbers suggest that acetate, one of the end products of anaerobic degradation of organic compounds, is an important substrate in this habitat. Unfortunately, no isolates were obtained from the zones from which the sequences were derived, and thus, it is not known whether bacteria related to Desulfobulbus spp. were present in the MPNs.
Another set of sequences (ML-9a-b, ML-16, ML-18, ML-20, and ML-21) formed a group that was affiliated with the Acidobacterium cluster (Fig. 2). All of the sequences derived from September samples (ML-16, ML-18, ML-20, and ML-21) formed a group of closely related sequences (between 96 and 97% identity), and they were more distantly related to the sequence derived from June samples (ML-9a-b) (93% identity). The taxon Acidobacterium has been considered a new kingdom (3) and at this time contains only three cultured representatives, but it also contains many uncultured forms that were identified by molecular analysis from a variety of environments (3). However, this is the first report of the occurrence of these organisms at a marine hydrothermal site. Inferences about their physiology cannot be made at present and must await cultivation of related bacteria.
The band ML-8 sequence was most closely related to clone OPB46 (94% sequence identity) (Fig. 2) belonging to the newly discovered candidate division OP9 (11). Members of this new line of descent in the domain Bacteria have so far been found only in a terrestrial hot spring, and the presence of related populations at a marine hydrothermal site extends the known habitat range for this group. The partial sequence in band ML-8 had a G+C content (58 mol%) similar to the G+C content of the corresponding region of OPB46 (59 mol%), for which a thermophilic nature has been inferred (11). Thus, it is likely that band ML-8 originated from a thermophilic population. This would be consistent with the finding that the band was present in the lower part of the DGGE gel, where it encountered a higher concentration of denaturant. Two other bands (ML-6 and ML-7) were also detected in the lower part of the DGGE gel and thus may also have originated from thermophilic populations. The fact that the closest relatives of the ML-7 organism were clones detected in a hot spring (11) supports this hypothesis (Fig. 2). However, as the level of identity was low (85%), placement of this sequence was uncertain. Clear thermophily could be inferred for the populations corresponding to ML-14a and ML-14b since the sequences in these bands were almost identical to the sequence of Caldicellullosiruptor lactoaceticus (level of sequence identity, almost 99%) (Fig. 2), which was isolated from a terrestrial hot spring (16). C. lactoaceticus is a thermophilic, cellulolytic, anaerobic bacterium that belongs to the Bacillus-Clostridium subphylum of low-G+C-content gram-positive bacteria (26). The finding that the sequences were obtained from water above the sediment at a distance of 30 cm seems inconsistent with a thermophilic and anaerobic physiology. However, this might have been related to resuspension of the upper sediment layers due to a storm before sampling (31). In deeper sediment layers the temperature increased rapidly and oxygen was absent (31; Ziebis et al., unpublished data).
Other populations (ML-4a-b, ML-6, and ML-17a-b) seemed to be only distantly related to previously described phylotypes (Fig. 2), and because we determined only partial sequences, their exact phylogenetic positions remain unknown. It is interesting, however, that bands ML-4a and ML-4b are affiliated with spirochetes, since the first anaerobic bacterium isolated from a deep-sea hydrothermal vent was a spirochete (10). Detection of a spirochetelike sequence by DGGE indicates that these organisms may play an important role in this environment.
Implications for trophic structure and similarities to other geothermal systems.
The data presented in this paper suggest that although autotrophic as well as heterotrophic populations could be detected, the microbial community at the vent site studied was predominantly heterotrophic. The potential substrates used by the microbial community are (i) organic matter produced at the vent site through photosynthesis by diatoms and through chemosynthesis by sulfur-oxidizing bacteria (e.g., Thiomicrospira spp.) and (ii) sea grass fragments from the surrounding sea grass meadows (1). This would provide an explanation for the presence of populations that degrade macromolecules, such as polysaccharides (e.g., C. lactocaceticus-related organisms and organisms that are affiliated with the CF phylogenetic branch). It has been suggested before that the occurrence of photosynthesis at this shallow-water vent site might lead to a phytodetritus-based food chain (35). This might be a major difference between this vent and most deep-sea hydrothermal vents, at which the input of allochthonous organic matter is low and the autochthonous organic matter is produced by chemosynthesis rather than by photosynthesis (12, 14). However, there might be similarities between the shallow-water vent which we studied and deep-sea vent sites with high rates of sedimentation of organic matter derived from the euphotic zone, such as Guaymas Basin (Mexico). At the latter site about 300 to 400 m of diatomaceous sediment that is rich in organic matter overlies the vents (33). This could lead to a higher proportion of heterotrophic organisms relative to the autotrophic populations (34), as observed at the vent which we examined.
There is only a limited database which can be used to compare the compositions of the microbial communities in different geothermal systems. An important similarity between the shallow-water vent site which we studied and deep-sea vents is that Thiomicrospira spp. are important components of both microbial communities (5, 20); populations related to the dominant phylotype at the shallow-water site (i.e., band ML-1) seem also to be present at deep-sea vents. It is noteworthy, however, that we did not detect any similarities between our results and the phylotypes that were found at an active deep-sea hydrothermal vent on Loihi seamount (18). On the other hand, there were similarities between the marine shallow-water hydrothermal vent and terrestrial hot springs in Yellowstone National Park (Obsidian Pool) and Iceland. Besides Obsidian Pool clone-related sequences and populations that are closely related to C. lactoaceticus, we also isolated a thermophilic sulfate-reducing bacterium (32) which is phylogenetically related to the Thermodesulforhabdus-Desulfacinum cluster in the delta subclass of the Proteobacteria. Populations belonging to this cluster were also found to be abundant in Obsidian Pool (11). Finally, it should be noted that this study included the first molecular analysis of a bacterial community at a marine shallow-water hydrothermal vent. Thus, it provides a framework with which to compare similar environments in the future.
Nucleotide sequence accession numbers.
The partial 16S ribosomal DNA sequences obtained in this study (DGGE bands ML-1a through ML-21) have been deposited in the GenBank nucleotide database under accession no. AF208985 through AF209019.
Acknowledgments
We are grateful to Wiebke Ziebis, Susanne Menger, and Guido Lützenkirchen for scuba diving, sampling, and assistance with the field work, to the mechanical workshop of the Max Planck Institute for Marine Microbiology for building the sampling devices, and to the participants of the EU-funded project Hydrothermal Fluxes and Biological Production in the Aegean for support and help and for an enjoyable stay on Milos. S.M.S. is indebted to Thorsten Brinkhoff for introducing him to DGGE analysis. We also thank Ferran Garcia-Pichel, Ulrich Nübel, Kerstin Sahm, and Hendrik Schäfer for helpful discussions and advice; Athena Econoumou-Amilli (Department of Biology, Section of Ecology and Systematics, University of Athens, Athens, Greece) for permission to cite unpublished data; and Geoffrey Mattison for linguistic improvements to the manuscript. We also acknowledge the Greek authorities for permission to undertake scuba diving and field work. Two anonymous referees provided valuable comments that improved the manuscript.
This work was funded by grant MAST CT-95-0021 from the EU and by the Max Planck Society, Munich, Germany.
REFERENCES
- 1.Aliani S, Bianchi C N, Cocito S, Dando P R, Meloni R, Morri C, Niemeyer A, Peirano A, Ziebis W. A map of sea grass meadows in Palaeochori Bay (Milos Island, Greece), a marine area with hydrothermal activity. Rapp Comm Int Mer Medit. 1998;35:512–513. [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns S M, Takala S L, Kuske C R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt K K, Ingvorsen K. Desulfobacter halotolerans sp. nov., a halotolerant acetate-oxidizing sulfate-reducing bacterium isolated from sediments of Great Salt Lake, Utah. Syst Appl Microbiol. 1997;20:366–373. [Google Scholar]
- 5.Brinkhoff T, Sievert S M, Kuever J, Muyzer G. Distribution and diversity of Thiomicrospira spp. at a shallow-water hydrothermal vent in the Aegean Sea (Milos, Greece) Appl Environ Microbiol. 1999;65:3843–3849. doi: 10.1128/aem.65.9.3843-3849.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dando P R, Aliani S, Bianchi C N, Cocito S, Fowler S W, Gundersen J, Hooper L, Kölbl R, Kuever J, Linke P, Makropoulos K C, Meloni R, Miquel J-C, Morri C, Müller S, Robinson C R, Schlesner H, Sievert S, Stöhr R, Stüben D, Thomm M, Varnavas S P, Ziebis W. Hydrothermal studies in the Aegean Sea. Phys Chem Earth B. 2000;25:1–8. [Google Scholar]
- 7.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel elctrophoresis profiles of 16S rRNA-defined populations inhabating a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox G E, Wisotzkey J D, Jurtshuk P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 9.Harmsen H J M, Prieur D, Jeanthon C. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl Environ Microbiol. 1997;63:2876–2883. doi: 10.1128/aem.63.7.2876-2883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harwood C S, Jannasch H W, Canale-Parola E. Anaerobic spirochete from a deep-sea hydrothermal vent. Appl Environ Microbiol. 1982;44:234–237. doi: 10.1128/aem.44.1.234-237.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jannasch H W, Mottl M J. Geomicrobiology of deep-sea hydrothermal vents. Science. 1985;229:717–725. doi: 10.1126/science.229.4715.717. [DOI] [PubMed] [Google Scholar]
- 13.Jannasch H W, Wirsen C O, Nelson D C, Robertson L A. Thiomicrospira crunogena sp. nov., a colorless, sulfur-oxidizing bacterium from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1985;35:422–424. [Google Scholar]
- 14.Karl D M. Ecology of free-living, hydrothermal vent microbial communities. In: Karl D M, editor. Microbiology of deep-sea hydrothermal vents. Boca Raton, Fla: CRC Press; 1995. pp. 35–124. [Google Scholar]
- 15.Llobet-Brossa E, Rossello-Mora R, Amann R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microb. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mladenovska Z, Mathrani I M, Ahring B K. Isolation and chracterization of Caldicellulosiruptor lactoaceticus sp. nov., an extremely thermophilic, cellulolytic, anaerobic bacterium. Arch Microbiol. 1995;163:223–230. [Google Scholar]
- 17.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek. 1998;73:127–141. doi: 10.1023/a:1000669317571. [DOI] [PubMed] [Google Scholar]
- 20.Muyzer G, Teske A, Wirsen C O, Jannasch H W. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch Microbiol. 1995;164:165–172. doi: 10.1007/BF02529967. [DOI] [PubMed] [Google Scholar]
- 21.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nübel U, Garcia-Pichel F, Kühl M, Muyzer G. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl Environ Microbiol. 1999;65:422–430. doi: 10.1128/aem.65.2.422-430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nübel U, Gracia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 25.Polz M F, Cavanaugh C. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc Natl Acad Sci USA. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rainey F A, Donnison A M, Janssen P H, Saul D, Rodrigo A, Bergquist P L, Daniel R M, Stackebrandt E, Morgan H W. Description of Caldicellulosiruptor saccharolyticus, gen. nov., sp. nov.: an obligately anaerobic, extremely thermophilic, cellulolytic bacterium. FEMS Microbiol Lett. 1994;120:263–266. doi: 10.1111/j.1574-6968.1994.tb07043.x. [DOI] [PubMed] [Google Scholar]
- 27.Reichenbach H, Dworkin M. The order Cytophagales. In: Balows A, Trüper H F, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. Vol. 4. Berlin, Germany: Springer-Verlag KG; 1992. pp. 3631–3687. [Google Scholar]
- 28.Rossello-Mora R, Thamdrup B, Schäfer H, Weller R, Amann R. The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Syst Appl Microbiol. 1999;22:237–248. doi: 10.1016/S0723-2020(99)80071-X. [DOI] [PubMed] [Google Scholar]
- 29.Round F E, Crawford R M, Mann D G. The diatoms. Biology and morphology of the genera. Cambridge, United Kingdom: Cambridge University Press; 1990. [Google Scholar]
- 30.Sievert S M. Microbial communities at a shallow submarine hydrothermal vent in the Aegean Sea (Milos, Greece). Ph.D. thesis. Bremen, Germany: Universität Bremen; 1999. [Google Scholar]
- 31.Sievert S M, Brinkhoff T, Muyzer G, Ziebis W, Kuever J. Spatial heterogeneity of bacterial populations along an environmental gradient at a shallow submarine hydrothermal vent near Milos Island (Greece) Appl Environ Microbiol. 1999;65:3834–3842. doi: 10.1128/aem.65.9.3834-3842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sievert S M, Kuever J. Desulfacinum hydrothermale, sp. nov., a thermophilic sulfate-reducing bacterium from geothermally heated sediments near Milos Island (Greece) Int J Syst Evol Microbiol. 2000;50:1239–1246. doi: 10.1099/00207713-50-3-1239. [DOI] [PubMed] [Google Scholar]
- 33.Simoneit B R T, Lonsdale P F. Hydrothermal petroleum in mineralized mounds at the seabed of Guaymas Basin. Nature. 1982;295:198–200. [Google Scholar]
- 34.Simoneit B R T. Hydrothermal petroleum: composition and utility as biogenic carbon source. Bull Biol Soc Wash. 1985;6:49–56. [Google Scholar]
- 35.Thiermann F, Akoumianaki I, Hughes J A, Giere O. Benthic fauna of a shallow-water gaseohydrothermal vent area in the Aegean Sea (Milos, Greece) Mar Biol. 1997;128:149–159. [Google Scholar]
- 36.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. In: Marshall K C, editor. Advances in microbial ecology. Vol. 12. New York, N.Y: Plenum Press; 1992. pp. 219–286. [Google Scholar]
- 37.Wenzhöfer F, Holby O, Glud R N, Nielsen H K, Gundersen J K. In situ microsensor studies of a shallow water hydrothermal vent at Milos, Greece. Mar Chem. 2000;69:43–54. [Google Scholar]
- 38.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer-Verlag KG; 1992. pp. 3352–3378. [Google Scholar]
- 39.Wirsen C O, Brinkhoff T, Kuever J, Muyzer G, Molyneaux S, Jannasch H W. A new Thiomicrospira strain from the Mid-Atlantic Ridge compared to known hydrothermal vent isolates. Appl Environ Microbiol. 1998;64:4057–4059. doi: 10.1128/aem.64.10.4057-4059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]