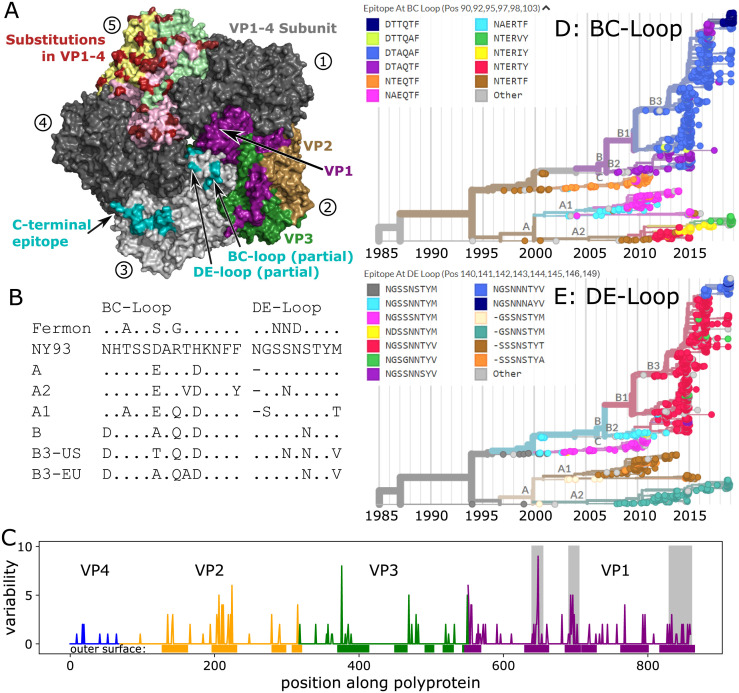
Fig 4. Molecular evolution of the EV-D68 capsid.
Panel A) Rendering of 5-fold symmetric arrangement of the crystal structure of the capsid protomer consisting of VP1, VP2, VP3 and VP4 [41], using different copies of the protomer to highlight different aspects of the capsid organization, evolution, and immunogenicity. The five subunits are labelled 1–5 (circled numbers). Subunits 1 and 4 are present in dark grey to complete the structure. Subunit 2 shows the surface exposed proteins in purple (VP1), sand (VP2), and green (VP3), subunit 3 displays the different putative epitopes [42] (note that the most variable parts of the BC- and DE-loops are not shown, as their structure is not resolved), and subunit 5 highlights variable positions in red, with the different proteins forming the surface indicated by pale colors. Panel B) The hypervariable BC- and DE-loops (partly missing from the structure in Panel A) in clusters and subclusters accumulated multiple changes since the root of the tree (the inferred root sequence patterns match those of the NY93 strain shown here). (BC-loop is AA positions 90–103; DE-loop is AA positions 140–148). Patterns for the C-terminus and a region of VP2 are shown in S2 Table. Panel C) Variable positions often fall into surface exposed parts of the protein (Fisher exact test, OR = 6, p < 10−16). Genes are highlighted by blue (VP4), orange (VP2), green (VP3), and purple (VP1). Grey boxes show, from left to right, the BC- and DE-loops, and C-terminus regions. Panels D&E) Phylogenetic VP1 trees are colored by the most common epitope patterns of the 6 and 8 most variable amino-acid positions in the BC- and DE-loops, respectively. Particularly note-worthy is the rapid turnover of BC-loop variants in the recent evolution of the A2 subclade. Patterns for the C-terminus region are shown in S6 Fig. See S8 Fig for a zoomed version of panels D & E with cumulative AA changes.