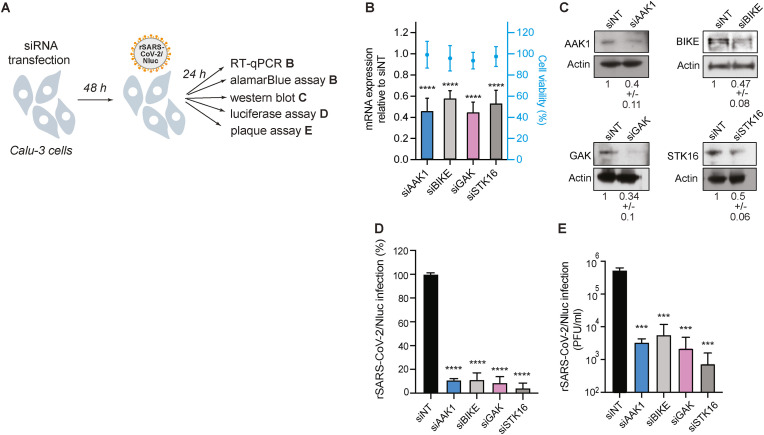
Fig. 1.
NAKs are required for SARS-CoV-2 infection.
(A) Schematic of the experiments shown in panels B–E. (B) Confirmation of siRNA-mediated gene expression knockdown and cell viability in Calu-3 cells transfected with the indicated siRNAs. Shown is gene expression normalized to GAPDH and expressed relative to the respective gene level in the non-targeting (siNT) control at 48 h post-transfection measured by RT-qPCR and cell viability measured by alamarBlue assay. (C) Confirmation of siRNA-mediated protein expression knockdown efficiency by western blot. Representative membranes and quantitative data from two experiments are shown. (D) WT SARS-CoV-2 infection measured at 24 h post-infection (hpi) of the indicated NAK-depleted Calu-3 cells with rSARS-CoV-2/Nluc (USA-WA1/2020 strain; MOI = 0.05) by Nluc assays. (E) Viral titers measured at 24 hpi of the indicated NAK-depleted Calu-3 cells with rSARS-CoV-2/Nluc (USA-WA1/2020 strain; MOI = 0.05) by plaque assays. Data in all panels are representative of two or more independent experiments. Individual experiments had three biological replicates, means ± standard deviation (SD) are shown. Data are relative to siNT (B, D and E). ***P < 0.001; ****P < 0.0001 by 1-way ANOVA followed by Dunnett's multiple comparisons test. PFU, plaque-forming units.