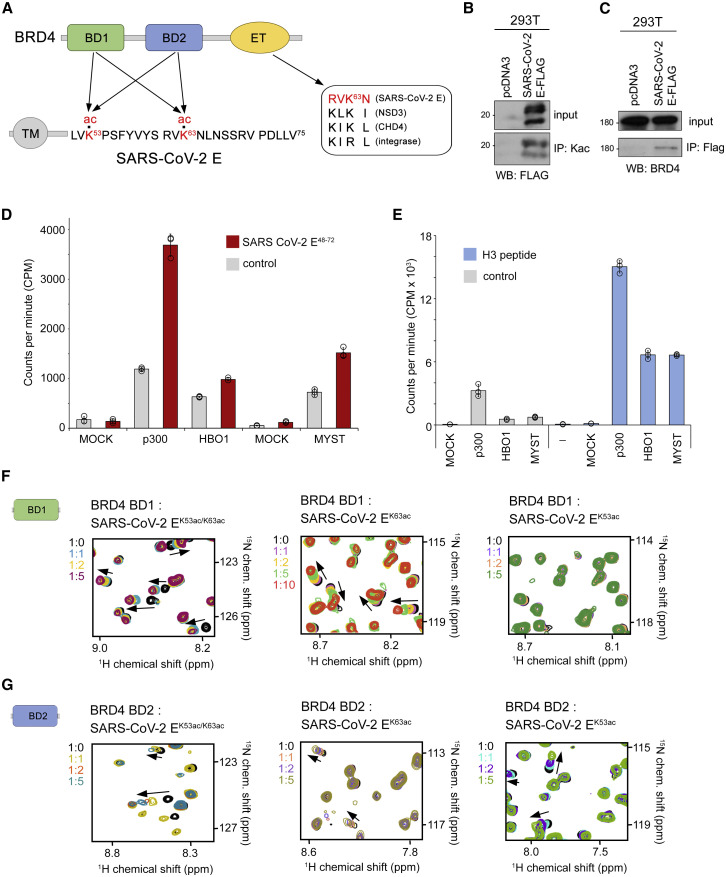
Figure 1.
SARS-CoV-2 E protein is acetylated in vivo and interacts with BRD4
(A) Domain architecture of BRD4 (top) and the SARS-CoV-2 E protein (bottom). Sequence alignment of the motif present in the E protein and other proteins known to interact with the BRD4 ET domain is shown on the right. Arrows indicate potential contacts between BRD4 and the E protein. Acetylated lysine residues are highlighted red.
(B) Immunoprecipitation with anti-acetyl-lysine antibodies on whole cell extracts from 293T cells expressing FLAG-tagged SARS-CoV-2 E protein or empty tag (pcDNA3) followed by western blot with FLAG antibody indicates that SARS-CoV-2 E is acetylated in vivo.
(C) Immunoprecipitation with anti-FLAG beads on the same extracts as in (B) followed by western blot with BRD4 antibodies indicates that the SARS-CoV-2 E protein interacts with BRD4 in vivo.
(D) Acetylation efficiency of the SARS-CoV-2 E protein in vitro. Acetyltransferase assays using recombinant p300 and the native human MYST-family HAT complexes purified via MEAF6 (MYST) or BRPF2 (HBO1) subunits and the SARS-CoV-2 E48-72 peptide (aa 48–72 of SARS-CoV-2 E) as the substrate. Incorporation of 3H-ac was measured by liquid scintillation counting. Mock purification from K562 cells expressing an empty tag was used as control. Data are represented as mean ± SD among three replicates.
(E) Acetyltransferase assays using recombinant p300 and H3 peptide (aa 1–29 of H3). Mock corresponds to a mock purification control. Data are represented as mean ± SD among three technical replicates.
(F and G) Overlayed 1H,15N HSQC spectra of 15N-labeled BRD4 BD1 (F) and BRD4 BD2 (G) recorded before and after gradual addition of the indicated acetylated SARS-CoV-2 E peptides. The spectra are color-coded according to the protein:peptide molar ratio. See also Figures S1, S2, and S4.